Optimization of Minimal Inhibitory Dose of Selective Agent (Basta) for Selection of Transgenics in Sugarcane 
Centre of Agricultural Biochemistry and Biotechnology (CABB), University of Agriculture Faisalabad, Pakistan.
Centre of Agricultural Biochemistry and Biotechnology (CABB), University of Agriculture Faisalabad, Pakistan
Centre of Agricultural Biochemistry and Biotechnology (CABB), University of Agriculture Faisalabad, Pakistan.
 Author
Author  Correspondence author
Correspondence author
Molecular Plant Breeding, 2012, Vol. 3, No. 5 doi: 10.5376/mpb.2012.03.0005
Received: 13 Mar., 2012 Accepted: 16 Apr., 2012 Published: 30 Apr., 2012
Ijaz et al., 2012, Optimization of Minimal Inhibitory Dose of Selective Agent (Basta) for Selection of Transgenics in Sugarcane, Molecular Plant Breeding, Vol.3, No.5 50-56 (doi: 10.5376/mpb.2012.03.0005)
Success rate in the selection of putative transformants is mainly dependent upon the dose rate of selective agent. Selection pressure determines the survival frequency of transformant and non transformant cells. Dose of selective agent, which is lethal for the non transformants, would be the optimum for the transgenic selection. We investigated the optimum dose of basta (selective agent), in two sugarcane genotypes viz., S-2003-us-359 and S-2003-us-127, in which we have already established a proficient in vitro regeneration system (Ijaz et al., 2012; Anjum et al., 2012). Different dosages of basta viz., 1 mg/L, 3 mg/L, 5 mg/L, 7 mg/L and 10 mg/L were studied for regeneration media (RM) of both genotypes. For determining the optimal inhibitory dose of basta on regeneration, 21 days old calli of S-2003-us-359 were induced on CIM1, 28 days old calli of S-2003-us-127 were induced on CIM2 and were shifted to RSM1 and RSM2 respectively. In genotype S-2003-us-359, early callus death was observed on RSM having 10 mg/L basta, and RSM having 5 mg/L, 7 mg/L, 10 mg/L basta proved to be lethal for S-2003-us-127 with early death of calli. For genotype S-2003-us-127, among 5 mg/L, 7 mg/L and 10 mg/L basta levels, 5 mg/L basta was selected for regeneration media as optimal dose for the selection of transgenics of this genotype. For transgenic selection in both sugarcane genotypes, the affectivity of selected optimal doses of basta was checked. Twenty one (21) days old calli of S-2003-us-359 and 28 days old calli of S-2003-us-127 were bombarded with bar gene and plant were selected at 10 mg/L and 5 mg/L of basta respectively. Putative transgenic plants with integrated copy of bar gene were confirmed with PCR analysis.
Transformation of crop genomes is used for many purposes such as increased nutritive value, plant disease resistance, insect resistance, herbicide resistance, and for high yield. The success of the genetic transformation process is monitored through the following 3 steps: the proof for DNA integration, protein expression and transmission of the transgene into its progenies. Practically, during genetic transformation, foreign gene is transferred into target tissues, which contains thousands of cells, and only a few cells will become transgenic or will have the transgene stably integrated into its genome. It is very important to isolate these transformed cells from the majority of untransformed cells by using a selection agent. The transform cells should carry a selectable marker gene, which will make the cell survive on a particular selection agent. It was reported that there are approximately 50 selectable marker genes being used or being developed in transgenic plant research (Miki and McHugh, 2004). In genetic transformation study the bar gene has been widely used as an effective selectable marker in many crop species (Gordon-Kamm et al., 1990; Akama et al., 1995). Bar gene was isolated from Streptomyces hygroscopicus (Block et al., 1987) and confers resistance to phosphinothricin, the active ingredient for the herbicides bialaphos, Liberty and Basta, an analogue of glutamate, a competitive inhibitor of the enzyme glutamine synthetase. Phosphinothricin acetyltransferase, coded by bar gene inactivates phosphinothricin by acetylation.
The concentration of selection agents need to be carefully chosen to avoid either being too low and thereby allowing undesirable numbers of ‘escapes’ to develop, or too high so that transformants expressing moderated levels of resistance are lost. Therefore optimization of selective agent is very crucial for selection of transgenic. Therefore, economical optimal concentration of the selective agent is the most preferred in the selection of stable transformed cells.
Selectable marker gene presence in transformed cells is necessary for the survival of the transformed cells on the selective agent. Until now almost 50 selectable marker genes are used in transgenic development research (Miki and Mchugh, 2004). Selective agent selection varies from crop to crop. Hygromycin as selective agent did not prove beneficial on the turf grass mature embryo development (Cao et al., 2006) while in case of apricot, kanamycin was proved to be excellent selective agent because it also improved the proliferation rate of the transformed tissues (Petri et al., 2005).
Basta is an efficient selective agent for monocot like sugarcane (Chowdhury and Vasil, 1992), rice (Christou et al., 1991), wheat (Vasil et al., 1992) and maize (Fromm et al., 1990). Therefore for our study basta was selected as selective agent for the selection of transgenics in sugarcane. So, Basta dose was optimized for sugarcane genotypes which were selected for genetic transformation study.
1 Results
The concentration of selection agent in the tissue culture media is very important. Less amount of selective agent may give rise to many false positives. Therefore an optimal amount of selective agent in the media is too necessary. Selection pressure determines the survival frequency of transformants and non transformed cells. Optimization of basta dose for genotypes viz., S-2003-us-359 and S-2003-us-127, five different concentrations of Basta (1, 3, 5, 7, 10 mgl ) were studied for transgenic selection on regeneration media. Basta concentrations for both genotypes were found which becomes lethal for non transformed cells. It was assumed that this concentration will not be lethal for transformed cells.
1.1 Selection of optimal Basta dose for genotype S-2003-us-359 to be used in regeneration selection media for transformation experiment
For the selection of optimal basta dose for regeneration media, calli were induced on CIM1 (Table 1). Twenty one (21) days old calli were transferred to a regime of Regeneration Selection Media (RSM) containing 1, 3, 5,7,10 mg/L basta (Table 2) respectively to select the optimal dose of basta, which was then used for the selection of transgenic plants from bombarded tissues. Calli on 10 mg/L basta showed death signs early. On second day of shifting, approximately 15% embryos died on this dose rate, and calli on 1,3,5,7 mg/L basta were unaffected. On the third day of shifting, approximately 55% embryos died on 10 mg/L and 10% embryos were died on 7 mg/L basta, but other calli were still unaffected. On 4th day, approximately 40%, 20%, and 10% death was observed on 5, 3, 1 mg/L basta respectively. More than 70 % death was observed on 10 mg/L basta and 50% embryos died on media having 7 mg/L basta. Fifth day, all calli died on me1dia containing 10 mg/L basta (Figure 2). But approximately 45%, 50%, 70% and 90% calli survived on 7, 5, 3 mg/L basta respectively (Figure 1; Figure 2).
Complete calli death, in case of 7, 5, 3 mg/L basta was observed on 9th, 11th and 14th day respectively. Up to 90% calli were survived on 1 mg/L basta even on 14th day. Though death started earlier on 10 mg/L basta but it was not so sudden and it seemed, on this basta dose if the calli are transgenic for bar they will survive and if not then die in 5 days.
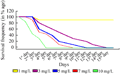 Figure 1 Optimization of basta dose for selection of transgenic regenerants in genotype S-2003-us-359; 10 mg/L basta showed early death and after 5 days on selection all calli were died. At 1 mg/L basta only 10% embryos died even after 15 days, and survival rate was 90% |
Figure 2 Calli response on Regeneration Selection Medium (RSM) at various levels of basta Note: A: 1 mg/L; B: 3 mg/L; C: 5mg/L; D: 7mg/L; E: 10 mg/L |
1.2 Selection of optimal Basta dose for genotype S-2003-us-127 to be used in regeneration selection media for transformation experiment
Calli of genotype S-2003-us-127 were induced on CIM2 (Table 1) and 28 days old healthy and proliferated calli were transferred on regeneration selection media (RSM) in which five levels of basta (1, 3, 5, 7, 10 mg/L) were used (Table 2) followed by incubation at 26±1℃ under 16/8 hrs light dark period. Proliferated calli were survived (100%) for one day only on all basta level. After one day of shifting on basta selection pressure, basta showed effect. It was observed that at 1 and 3 mg/L calli survived (80%) while at 5, 7 and 10 mg/L calli survival response was 50, 40 and 30% respectively. On fifth day, survival frequency of embryo was 5 and 3% on 5, 7, and 10 mg/L basta level respectively. While complete death of embryo was observed at 5, 7, and 10 mg/L basta level on 7th day (Figure 3 and Figure 4).
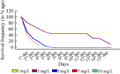 Figure 3 Optimization of basta dose for selection of transgenic regenerants in genotype S-2003-us-127 |
Figure 4 Calli response on Regeneration Selection Medium (RSM) at various levels of basta Note: A: 1 mg/L; B: 3 mg/L; C: 5mg/L; D: 7mg/L; E: 10 mg/L |
But 50% and 45 % embryo were survived at 1 and 3 mg/L basta respectively. On 1 mg/L basta regeneration was starting after 12 days (Figure 5).
After the selection of basta doses for genotype S-2003-us-359 and S-2003-us-127, affectivity or efficiency of these optimal doses was determined by integrating the bar gene in to these genotypes using gene gun DNA delivery method. Putative transgenic plants of both genotypes were selected on their respective optimal basta dose.
Figure 5 Regeneration on RSM having 1 mg/L basta |
1.3 Selection of transgenic plants of both genotypes
Twenty one (21) days old calli of genotype S-2003-us-359 and 28 days old calli of S-2003-us-127 those were induce on CIM1 and CIM2 respectively were bombarded with bar gene and shifted to RSM having 10 and 5 mg/L basta. RSM with 10 mg/L basta was used for the selection of transgenics of genotype S-2003-us-359 and RSM with 5 mg/L basta was used for the selection of transgenics of genotype S-2003-su-127 (Figure 6).
 Figure 6 Selection of transgenics on selection regime having basta as selective agent: (A & B) transgenic plants selection of genotype S-2003-us-359 on RSM having 10 mg/L basta; (C & D) transgenic plants selection of genotype S-2003-us-127 on RSM having 5 mg/L basta |
1.4 Genomic analysis of putative transgenic plants with PCR
Putative transgenic plants those bears the selection pressures of basta were confirmed for the presence of bar gene in their genome with PCR analysis by using gene specific primers. DNA of wild type plant as well as putative transgenics those bear basta selection pressure were extracted. PCR was performed by using gene specific primers. bar gene sequence was absent in wild type (non transformed plant) but presence of bar gene sequence was observed in putative transgenics (transformed plants) those were selected by giving the selection pressure of basta (Figure 7).
 Figure 7 PCR analysis of putative transgenics for bar; M = I kb ladder, E= Empty lane, WT= Wild type plants, P1, P2, P3 =Transgenic plants of genotype S-2003-us-359, P4, P5 =Transgenic plants of genotype S-2003-us-127, -ve control (water), E=Empty lane, + ve control (plasmid DNA) |
2 Discussion
Selection of transformed cells containing stably integrated gene is one of the major steps in production of transgenic plants which can be achieved by knowing the minimum concentration of selective agent that can inhibit the growth of non-transformed cells and allow transformed cells to survive. Due to this, transformation process has become more efficient that results in a very low occurrence of chimeras. Sreeramanan et al., (2006) highlight the significance of dose rate optimization of the selective agent and determine that it is the most important step in the selection of the transformed plants because it makes the selection process very easy and efficient. Dose rate optimization of the selective agent is highly tissue and species specific described by Parveez et al (1996). Due to high dependence against genotype in monocot it might be the variation in the endogenous resistance. Therefore, optimization of the selective agent becomes very crucial. Under these circumstances, this study was organized for the efficient selection of transgenic plants. It was observed that basta is an efficient selective agent in the selection of sugarcane transformed plants. It was also reported that prolonged exposure to selection in medium containing Basta is needed to reduce escapes.
Basta dose was optimized for transgenic selection and five levels of basta (1 mg/L, 3mg/L, 5 mg/L 7 mg/L and 10 mg/L) were used. 10 mg/L basta was selected for transformation study of genotype S-2003-us-359 and 5 mg/L basta was selected for the transformation study of S-2003-us-127 because at these level early calli death was observed but Ahmad (2009) used 3 mg/L basta dose for transgenic selection in wheat. Manickavasagam et al (2004) suggested 5 mg/L basta as most effective selective agent for transgenic selection in sugarcane. These results showed, the dose of selective agent (basta) varying in both genotypes and these results reveal that dose rate of selective agents is also genotype dependent. These results were also agreed with the results of van Boxtel et al (1995), who also reported that the sensitivity to selective agents was genotype dependent. Therefore it concluded that optimization of optimal dose of selective agents should be done before the transformation of any crop plant.
3 Materials and methods
3.1 Germplasm/ plant materials
Two genotypes were selected viz., S-2003-us-359 and S-2003-us-127, in which we have already established an in vitro regeneration system (Ijaz et al., 2012; Anjum et al., 2012).
3.2 Selection of basta dose rate for in vitro regeneration media to select transgenics
Young innermost leaves of sugarcane genotypes viz., S-2003-us-359 and S-2003-us-127 were cultured on CIM 1 and CIM 2 respectively (Table 1). For determine the minimal inhibitory dose of basta on regeneration, Calli of both genotypes were induced on their respective callus induction media (CIM).
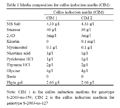 Table 1 Media composition for callus induction media (CIM) |
Twenty one (21) days old Calli of S-2003-us-359 and 28 days old calli of S-2003-us-127 were transferred to regeneration selection medium 1 (RSM 1) and regeneration selection medium 2 (RSM 2) respectively. RSM 1 containing 2, 4-D (0.1mg/L), BAP (0.25 mg/L), sucrose (40 g/L) and basal MS Salt with different levels (1, 3, 5, 7, and 10 mg/L) of basta (selective agent) but RSM 2 containing 2, 4-D (0.1 mg/L), BAP (1 mg/L), sucrose (30 g/L) and basal MS Salt with different levels (1, 3, 5, 7, and 10 mg/L) of basta (Table 2). Experiment was repeated three times and data were collected on the basis of survival rate.
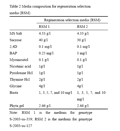 Table 2 Media composition for regeneration selection media (RSM) |
3.3 Genetic transformation of both genotypes with bar gene to check the efficiency of selected basta doses for transgenic selection
Genotype S-2003-us-359 and S-2003-us-127 were genetically transformed with bar gene by using gene gun delivery method to check the affectivity of selected doses of basta for transgenic selection in both genotypes. Transgenic plants were selected on selection regime having at 10 mg/L basta and 5 mg/L basta for genotype S-2003-us-359 and S-2003-us-127 respectively.
3.4 Preparation of gold particle
Gold particles (40 mg) with an average size (0.6 µM) were suspended in 1 ml of 96% ethanol. Centrifugation was done at 4 200 rpm for 1 minute. Supernatant was removed followed by the addition of 1 ml (96%) ethanol. Resuspended the particles again for short time and repeated three times. Particles were washed in 1 ml ultra pure water for three times. Resuspended the particles again in 1ml ultra pure water after last centrifugation treatment. Aliquots of 50 µl were formed and stored aliquots at -80℃.
3.5 Bombardment of calli with bar gene and selection of putative transformed plants
The plasmid DNA having bar gene (Figure 8) was precipitated on to 0.6 μ gold particles. DNA coated gold particles were bombarded (PDS 1000/He). The bombarded calli of genotype S-2003-us-359 and S-2003-us-127 were transferred to Regeneration selection media having 5 and 10 mg/L basta respectively, incubated for 8~16 hrs dark and light condition at 26±1℃. Only those plants were survived on basta which have bar gene and others were died.
Figure 8 PCa bar Expression cassette with 35S promoter, bar and T35S terminator and restriction sites |
3.6 DNA extraction
Leaf tissues (200 mg) were ground in liquid nitrogen 700 µL extraction buffer was added in each reaction tube followed by the addition of 800 µL phenol chloroform Isoamylalcohol (25:24:1). Centrifugation was done for 3 minute at 5 000 rpm at 4℃. Supernatant was taken followed by the addition of 1/10 of sodium acetate into each tube. Equal volume of Iso-propanol was added into each tube. Centrifugation was performed at 13 200 rpm for 15 minutes. Supernatant was removed. Washing of pellets with 80% ethanol was done, followed by air drying the pellets. The pellet was dissolved in R40 (40 μg/ml RNAse A in 1x TE, pH 8.2). DNA quantity and quality was checked with nanophotometer and by running on 0.8% agarose gel.
3.7 PCR analysis
Plants which were survived on basta were analyzed with PCR (Polymerase chain reaction) analysis. Putative plants which were survived on basta were compared with wild type. PCR was carried out in a 25 µl reaction volume, containing 13.75 µl d3H2O, 2.5 µl 10×Taq buffer, 2.5 µl MgCl2, 1 µl dNTPs, 0.25 µl Taq DNA Polymerase, 1 µl of each primer (reverse and forward primers flanking the bar gene) and 3 µl Template DNA. The sequences of reverse and forward primers of bar gene are as follows.
Table 3 Primers used for the amplification of bar gene for transgenic confirmation |
3.8 PCR (RAPD) amplification profile
PCR amplification was done by incubating the each DNA samples at 95℃ for 3 minutes, then 35 cycles comprising of denaturation at 95℃ for 1 minute, annealing of primers at 58℃ for 1 minute and extension at 72℃ for 1 minute. The final extension was carried out at 72℃ for 10 minutes. Resolving of PCR product was done by gel electrophoresis using ethidium bromide staining solution. Agarose gel electrophoresis separates macromolecules on the basis of charge, size, or other physical properties. PCR product were resolved on 0.8 Agarose in 0.5×TAE. After electrophoresis, gels were photographed using gel documentation system, and gel pictures were saved.
Reference
Akama K., Puchata H., and Hohn B., 1995, Efficient Agrobacterium mediated transformation of Arabidopsis thaliana using the bar gene as selectable marker, Plant Cell Rep., 14: 450-454 http://dx.doi.org/10.1007/BF00234053
Anjum N., Ijaz S., Rana I.A., Khan T.M., Khan I.A., Khan M.N., Mustafa G., Joyia F.H., and Iqbal A., 2012, Establishment of an in vitro regeneration system as a milestone for genetic transformation of sugarcane (Saccharum officinarum L.) against Ustilago scitaminea, Bioscience Methods, 3(2): 7-20
Cao M. X., Huang J.Q., He Y.L., Liu S.J., Wang C.L., Jiang W.Z., and Wei Z.M., 2006, Transformation of recalcitrant turf grass cultivars through improvement of tissue culture and selection regime, Plant Cell Tissue and Organ Culture, 85: 307-316 http://dx.doi.org/10.1007/s11240-006-9081-7
Chowdhury M.K.U., and Vasil I.K., 1992, Stably transformed herbicide resistance callus of sugarcane via micropro¬jectile bombardment of cell suspension cultures and electroporation of protoplasts, Plant Cell Reports, 11: 494-498 http://dx.doi.org/10.1007/BF00236264
Christou P., Ford T., and Kofron M., 1991, Production of transgenic rice (Oryza sativa L.) plants from agronomi¬cally important indica and japonica varieties via electric discharge particle acceleration of exogenous DNA into immature zygotic embryos, Bio/Technology, 9: 957-962 http://dx.doi.org/10.1038/nbt1091-957
Block, D.M., Botterman J., Vandewiele M., Dockx J., Thoen C., Gossele V., Rao V., Movva N., Thompson C., Montagu M.V., and Leemans J., 1987, Engineering herbicide resistance in plants by expression of a detoxifying enzyme, EMBO J., 6: 2513-2518 PMid:16453789 PMCid:553667
Fromm M.E., Morrish F., Armstrong C., Williams R., Thom¬as J., and Klein T.M., 1990, Inheritance and expression of chimeric genes in the progeny of transgenic maize plants, Bio/Technology, 8: 833-838
http://dx.doi.org/10.1038/nbt0990-833 PMid:1366794
Gordon-Kamm W., Spencer T.M., Mangano M.L., Adams T.R., Daines W.G., Start J.V., O'Brien S.A., Chambers W.R.J., Adams N.G., Willetts N.G, Rice T.B., Mackey C.J., Krueger R.W., Kausch A.P., and Lemaux P.G., 1990, Transformation of maize cells and regeneration of fertile transgenic plants, The Plant Cell, 2: 603-618
http://dx.doi.org/10.2307/3869124 PMid:12354967 PMCid:159915 http://dx.doi.org/10.1105/tpc.2.7.603 PMid:12354967
Ijaz S., Rana I.A., Khan I.A., and Saleem M., 2012, Establishment of an in vitro regeneration system for genetic transformation of selected sugarcane genotypes, Genet. Mol. Res., 11 (1): 512-530
http://dx.doi.org/10.4238/2012.March.6.4
Manickavasagam M., Ganapathi A., Anbazhagan V.R., Sudhakar B., Selvaraj N., Vasudevan A., and Kasthurirengan S., 2004, Agrobacterium-mediated genetic transformation and development of herbicide-resistant sugarcane (Saccharum species hybrids) using axillary buds, Plant Cell Rep., 23: 134-143
http://dx.doi.org/10.1007/s00299-004-0794-y PMid:15133712
Miki B., and McHugh S., 2004, Selectable marker genes in transgenic plants: applications, alternatives and biosafety, Journal of Biotechnology, 107: 193-232 http://dx.doi.org/10.1016/j.jbiotec.2003.10.011 PMid:14736458
Parveez G.K.A., Chowdhury M.K.U., and Saleh N.M., 1996, Determination of minimal inhibitory concentration of selection agents for oil palm (Elaeis guineensis Jacq.) transformation, Asia Pacific J. Mol. Biol. Biotech., 4: 219-228
Petri C., Albuquerque N., and Burgos L., 2005, The effect of aminoglycoside antibiotics on the adventitious regeneration from apricot leaves and selection of nptII-transformed leaf tissues, Plant Cell, Tissue and Organ Culture, 80: 271-276 http://dx.doi.org/10.1007/s11240-004-1019-3
Rana A. I., 2009, Reverse and Forward Genetic Approaches for the Development of Disease Resistant Wheat (Triticum aestivum L.), PhD thesis, University of Agriculture Faisalabad, Pakistan
Sreeramanan M., Maziah M., Abdullah M.P., Rosli N.M., and Xavier R., 2006, Potential selectable marker for genetic transformation in Banana, Biotech., 5: 189-197 http://dx.doi.org/10.3923/biotech.2006.189.197
Vasil V., Castillo A.M., Fromm E.M., and Vasil I.K., 1992, Herbicide resistant transgenic wheat plants obtained by microprojectile bombardment of regenerable embryo¬genic callus, Bio/Technology, 10: 667-674
http://dx.doi.org/10.1038/nbt0692-667
Van Boxtel J., Eskes A., and Berthouly M., 1995, Glufosinate as an efficient inhibitor of callus proliferation in coffee tissue, In Vitro Cell. Dev. Biol. Plant, 33: 6-12 http://dx.doi.org/10.1007/s11627-997-0033-7
. PDF(1291KB)
. FPDF
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Siddra Ijaz
. Naweed Anjum
. Iqrar Ahmad Rana
. Iqrar Ahmad Khan
Related articles
. Basta
. Bar gene
. Sugarcane
. Transgenics
Tools
. Email to a friend
. Post a comment


