Ectopic expression of an AGAMOUS homolog NTAG1 from Chinese narcissus accelerated earlier flowering and senescence in Arabidopsis 
 Author
Author  Correspondence author
Correspondence author
Molecular Plant Breeding, 2011, Vol. 2, No. 3 doi: 10.5376/mpb.2011.02.0003
Received: 28 Dec., 2010 Accepted: 06 Jan., 2011 Published: 29 Jan., 2011
Deng et al., 2011, Ectopic expression of an AGAMOUS homolog NTAG1 from Chinese narcissus accelerated earlier flowering and senescence in Arabidopsis, Molecular Plant Breeding Vol.2 No.3 (doi: 10.5376/mpb.2011.02.0003)
There are two cultivating varieties of Chinese narcissus, named as Yulinglong (Narcissus tazetta var. chinensis Roem. Florepleno) and Jinzhanyutai (Narcissus tazetta var. chinensis M. Roem.) well known in China. Yulinglong plants exhibit double flower resulted from petaloid stamens. However, the molecular basis of double flower formation is known little and unclear. Based on the flowering ABCDE model, double flower formation is commonly used to link to C functional genes. In this present paper, the isolation and characterization of NTAG1 gene, an AGAMOUS homolog from Chinese narcissus varieties mentioned above are reported. Sequence and expression pattern of NTAG1 gene exhibited the same in both tested varieties. It expresses only in the reproductive organs. Furthermore, functional analysis by using ectopic tests in Arabidopsis showed that NATG1 might be involved in the carpel identity and floral transition The effects of ectopic expression of NTAG1 mainly include dwarfing , early flowering, losing inflorescence indeterminacy,branch number increasing, and advancing senescence, whereas some of homeotic phenotypes gradually sisappeared in higher generation of transgenic plants. The utilizations of NATG1 gene in the future gene engineering were also discussed in the paper.
Background
Certain flower characters including the flowering time, floral architecture and petal color are usually concerned by ornamental plants breeders. Double flowers resulted from increased number of petals were selected for their showy appearance in many domesticated plant families. Chinese narcissus (Narcissus tazetta var. chinensis) is popular flower with high cultural value in the ornamental market.But the problem of unitary culture variety have become to be the restrict factor for the Chinese Narcissus industry achieving further development. There are only two cultivating varieties, named as Yulinglong (Narcissus tazetta var. chinensis Roem. Florepleno) and Jinzhanyutai (Narcissus tazetta var. chinensis M. Roem.) respectivelly, in Chinese narcissus. The flower of Yulinglong exhibits double flower resulted from petaloid stamens. However, the molecular basis of the double flower formation is known little and unclear. And there are only a few reports about flower formation of Chines narcissus.
The genetic mechanism regulating floral formation is extensively studied in model plants and the ABCDE model has been established (Bowman et al., 1989; Parcy et al., 1998; Pelaz et al., 2001; Pelaz et al., 2000; Roeder and Yanofsky, 2001). In this model, A and E gene classes determine sepal identity, A, B and E determine,petal, B, C and E specify stamen , C and E specify carpel and D functional gene is involved in the development of ovule identity. Functional flower organ identity genes including. APETALA1 (A class), APETALA3 and PISTILLATA (B class), AGAMOUS (AG) (C class), FLORAL-BINDING PROTEIN (FBP) 7 and FBP11 (D class), and SEPALLATA1/2/3 are ( E class) have been isolated (Coen and Meyerowitz, 1991; Colombo et al., 1997; Colombo et al., 1995; Drews et al., 1991; Jofuku et al., 1994; Mandel et al., 1992; Pelaz et al., 2000; Rounsley et al., 1995; Theissen, 2001; Weigel and Meyerowitz, 1993; Weigel et al., 1992). Most of these genes (except APETALA2) belong to MADS-box transcription factors gene family which control floral organ identity (Melzer et al., 2009; Saedler et al., 2001; Theissen, 2001). The C-function gene AG plays a central role not only in specifying sexual organ identity but also in determining floral meristem termination (Bowman et al., 1989; Lohmann and Weigel, 2002). In Arabidopsis, AG mutation results in the expansion region of the A gene class into the center of the flower, which makes stamens change into petals and carpels into sepals. In addition,there are additional abnormal flower produced in the center of ag flower. There is increasing evidence that the role of floral organ genes is conserved in different plants, although differences in regulation, redundancy and function of these genes exist between species (Ferrario et al., 2006).
Based on the flowering ABCDE model, double flower formation is commonly used to link to C functional genes. NTAG has been reported being a putative AG ortholog cloned from Chinese narcissus only by the sequence similarity and the expression pattern analysis (Wang et al., 2006). However, no further functional analysis was performed to indicate its involvement in carpel development. In this presrnt paper the isolation and characterization of NATG1 gene, an AG homolog from both Chinese narcissus varieties mentioned above are reported. Sequence and expression pattern of NTAG1 gene exhibited the same in both tested varieties. Furthermore, functional analysis by using ectopic tests in Arabidopsis showed that NATG1 might be involved in the carpel identity and floral transition. The utilizations of NATG1 gene in biotechnology are discussed.
1 Results
1.1 Isolation and characterization analysis of NTAG1 genes from two narcissus varieties plants
The flowers of Jinzhanyutai consist of five whorls of organs including three sepals, three petals, a golden cup-shaped corona, six stamens, and three fused carpels (Figure1A). The sepals and petals are white, extremely similar and known as tepals (Figure1A). Yulinglong plants produce metamorphotic flowers in which stamens are petal-like structure (Figure1B, C). In order to study the mechanism of such double flower development,we isolated the NTAG1 gene, a putative C-function gene with the information reported by Wang (Wang et al., 2006) from both narcissus plants respectively. Sequence analysis showed that NTAG1 genes from both varieties plants were exactly same. Comparison of the sequence with that presented by Wang (Wang et al., 2006) showed that there were 5 bases pair variation between them. However, there was only one residue changed in the K-box when the deduced amino acid sequence was compared (data not shown). The reason of the sequence difference may be caused by PCR or resulted from the plant materials got from different places.
|
Figure 1 A-C: The flower structure of Chinese narcissus, Bar=4 mm; A: The flower of Jinzhanyutai; B: The flower of Yulinglong; C: The innerest two wholes of Yulinglong; The arrow showes the carpel; D-H: Phenotypes of transgenic Arabidopsis plants expressing 35S::NTAG1 in the T1 generation; D: A 14-d-old Wt Arabidopsis (Ler), Bar=3 mm; E-F: A 14-d-old (E) and a 20-d-old (F) NTAG1 overexpressing plant in Ler showed reduced size, early flowering, and small and curled leaves; Arrow shows thin and curled leave with yellow tips, Bar=3 mm; G-H: Eighteen-d-old transgenic plants expressing 35S::NTAG1 in Col in the T1 generation. Wild type and transgenic phenotype plants segregated in this generation; All transgenic plants flowered significantly early; Some lines of transgenic plants showed significantly small structure (showed by arrows in G) with only 1 cm tall during this stage; Some lines showed similar tall as Wt but with obvious curled rosette leaves (H), Bar=3 mm; I: Most flowers of 35S::NTAG1 in Col in the T1 generation are smaller, Bar=250 μm; J: First-whorl organs were transformed into carpelloid sepals with stigmatic papillae (showed by arrows), Bar=2 mm; K: Compared with Wt, the 35S::NTAG1 transgenic plants flowered early (left) by producing only six-seven small, curled rosette leaves (right), Bar=5 mm/L; The 35S::NTAG1 transgenic plants (right) began to senescence when Wt plants (left) bolted, Bar=10 mm; M-N: Scanning electron micrographs of sepals observed in Wt (M) and 35S::NTAG1 transgenic plants (N), Bar=20 μm
|
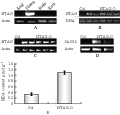
The NTAG expression has been shown only in the third and forth whorl of the flower of Jinzhanyutai by northern blot (Wang et al., 2006). To further explore whether its expression changed in Yulinglong, semi-quantitative RT-PCR was performed and the results revealed that the expression pattern was nearly similar in both varieties plants, it was only expressed in flowers, but not detectable in leaves, stems and roots (Figure2A).
|
Figure 2 A: The expression of NTAG in Chinese narcissus; B: The 35S::NTAG1 transgenic plants were verified by PCR using genomic DNA as templates; C: The 35S::NTAG1 transgenic plants were verified by RT- PCR; D: The expression SAG12 gene was enhanced in the 35S::NTAG1 transgenic plants; E: The MDA content was higher in the 35S::NTAG1 transgenic plants
|
1.2 Ectopic expression of NTAG1 caused early flowering and affected the floral organ identity in transgenic Arabidopsis plants
To further investigate whether the sequence and structure similarity (Wang et al., 2006) is coupled to the functional similarity between NTAG1 and C functional genes, We performed functional analysis through transgenic plants. NTAG1 cDNA driven by cauliflower mosaic virus 35S promoter was therefore transformed into Arabidopsis plants. Twenty-one independent transgenic Arabidopsis T1 plants in Col and nine lines in Ler ecotype were obtained through kanamycine screening (results not shown) and PCR (Figure 2B). Most transformed plants produced nearly similar and severe transformation in all development (Figure 1D-J). NTAG1 overexpressing plants in Ler showed reduced size (Figure1E, compared with 1D), early flowering, and small and curled leaves with yellow tips (Figure1F). Wild type and transgenic phenotype plants were segregated in T1 generation. All transgenic plants flowered significantly early. Some lines of transgenic plants showed significantly small structure (showed by arrows in Figure 1G). Some lines showed similar tall as Wt but with obvious curled rosette leaves (Figure 1H). The structure of most flowers of 35S::NTAG1 in Col and Ler in the T1 generation was similar as that of wild type (Wt), however the size of flowers was smaller (Figure 1I). Three lines of transgenic plants produced transformed flowers, first-whorl organs were transformed into carpelloid sepals containing stigmatic papillae (showed by arrows in Figure 1J) and second whorl organs absent sometimes, the typical homeotic conversion of sepals and petals similar to that observed in ap2 mutants or AG ortholog overexpressed transgenic Arabidopsis plants (Mizukami and Ma, 1992; Mizukami and Ma, 1997; Tzeng et al., 2002).
Because the phenotypes varied with the expression of the transgenic gene, the expression of NTAG1 gene was analyzed in five lines of the homozygous kanamycin-resistant T3 generation plants by RT-PCR. The results showed that NTAG1 gene were ectopic expressed in these plants (Figure 2C). Compared with Wt, the 35S::NTAG1 transgenic plants flowered early (Figure 1K, left) by producing only six-seven small, curled rosette leaves (Figure1K, right, Table 1), loss of inflorescence indeterminacy. The branch number and the number of inflorescence buds of the transgenic plants were increased (Table 1). All transgenic plants produced terminal flowers (Figure 1K) similar to those observed in Arabidopsis plants ectopically expressing the AG or AG orthologs (Mizukami and Ma, 1992; Rutledge et al., 1998). However, there was no obvious homeotic conversion of flower organs in the transgenic plants. When the epidermal cells of the flower organs were examined by scanning electron micrographs, the inner three whorls were morphologically similar to the Wt organs epidermis (data not shown). The surface of irregularly shaped cells in Wt sepals are cuticular thickening, whereas the surface of irregularly shaped cells along with the interspersed stomata in Wt carpels are smooth (Tzeng et al., 2002). A 35S::NTAG1 flower produced sepals nearly similar to wild type except that surface cells containing less cuticular thickenings (Figure 1M, N). Additionlly,we also investigated, the morphological features of transgenic plants (Table 1). The height of 35S::NTAG1 plants was 5.6 cm, which was shorter than that of Wt (11.5 cm). The leaf was smaller, and the silique was shorter (the average silique length of 35S::NTAG1 plants was 7.5 mm versus 10.5 mm of Wt), and the number of branch per plant increased significantly (the average branch number of 35S::NTAG1 plants was 8.1 versus 4.0 of Wt).
|
Table1 Morphological features of wild type and 35S::NTAG1 plants
|
1.3 Ectopic expression of NTAG1 accelerated senescence in transgenic Arabidopsis plants
Even in the vegetative stage, the leaves produced by the 35S::NTAG1 transgenic plants were often yellow which is characteristic of senescence. Most leaves of the transgenic plants began to lose green when Wt plant bolted (Figure 1L). To analyze the effect of the ectopic expression of NTAG1 on senescence, the MDA content, a stress or senescence indicator, was assayed in the 14-d-old plants. It showed that the transgenic plants had much higher MDA level than Wild type plants (Figure 2E). In addition, the expression of SENESCENCE ASSOCIATE GENE 12 (SAG 12), a senescence marker (Gan and Amasino, 1995) was advanced by ectopic expression of NTAG1 (Figure 2D).
2 Discussion
In Arabidopsis, AG is required for both floral meristem determinacy and reproductive organ identity (Bowman et al., 1989; Lohmann and Weigel, 2002). In Arabidopsis, AG mutation results in the expansion region of the A gene class into the center of the flower, which makes stamens change into petals and carpels into sepals. In addition,,there are additional abnormal flower produced in the center of ag flowers suggesting that the floral meristem of this mutant is indeterminate. To explore the possible molecular mechanisms involved in double Narcissus flower, we isolated and characterized NTAG1 gene, an AG homolog from both Chinese narcissus varieties. Both the results that the sequence and expression pattern exhibited same in both varieties, and evidence of functional NTAG1 gene existing in double flowers showed by ectopic tests in Arabidopsis, indicate that the increase of petal number in narcissus flower is not caused by the sequence mutation or a deregulation of expression of NTAG1 gene. Gao etc. al (Gao et al., 2008) isolated a MADS-box gene NTMADS1, an AG ortholog, from Chinese narcissus double flower plants and speculated that the sequence variation from that of NTAG is the reason of double flower formation in narcissus. According to our data, such sequence difference may be caused by PCR or resulted from the plant materials got from different places. Based on the flowering ABCE model, the double flower might be caused by the restriction of the AG ortholog expression, or by the expansion of A-function genes expression domains (Dubois et al., 2010). So further analysis of NTAG1 expression domains, the isolation and characterization of other C-function genes and A-function genes in narcissus, will be necessary to draw a clear conclusion.
Not similar to the most AG homolog transgenic plants (Mizukami and Ma, 1992; Mizukami and Ma, 1997; Tzeng et al., 2002), ectopic expression of NTAG1 in Arabidopsis is not often accompanying with the homeotic transformation flower (Less than 10% lines in T1 generation). Especially with the generation increased, the transgenic plants developed normal flowers. There are three explanations for such data. Firstly, the expression of NTAG1 gene in most transgenic plants, especially in higher generations, was not enough for the flower transformation. Secondly, NTAG1 gene might not be the only one C-function genes in narcissus. Indeed, the C-function is shared by two partially redundant genes in many plant species (Dubois et al., 2010). For instance, AG performs the sexual organ identity and floral meristem termination, whereas, SHATTERPROOF (SHP) is involved in later carpel development stages in Arabidopsis (Causier et al., 2005). Conversely, in Antirrhinum majus L., the ortholog of SHP, PLENA is essential for sexual organ identity (Davies et al., 1999). It is therefore of interest to identify SHP lineage in narcissus. Additionally, in line with the quartet model (Theissen, 2001), another reason might be the different NTAG1 interaction factors existed in narcissus from that in Arabidopsis.
The novel phenotypes of the NTAG1 transgenic plants include early flowering, losing inflorescence indeterminacy and increasing branch number, which are usually candidate characters of ornamental plants selected by people. So it will be an ideal candidate gene for the genetic modification in narcissus or other plants. However, ectopic expression of NTAG1 accelerated earlier senescence. This might be a consequence of the competition between sink and source, the poor source caused by fewer and smaller leaves, the competitive sink characterized by early reproductive development. So in order to obtain more novel phenotypes, adjust the NTAG1 expression driven by certain promoter such as inducible promoter await for future performance.
3 Materials and methods
3.1 Plant materials and growth conditions
Plants of narcissus Yulinglong and Jinzhanyutai used for this study were grown in the field in Chongming, Shanghai. Arabidopsis thaliana of ecotype Columbia-0 (Col) and Lersberg (Ler) plants were grown in the green house under constant illumination (-80 μ mol·m–2·s–1) at 22±2ºC. For transgenic plants to be screened, seeds were surface sterilized, stratified at 4ºC for 3 d, plated on 1/2 Murashige and Skoog (Murashige and Skoog, 1962) medium (MS) containing 50 μg·mL-1 kanamycin, and grown under continuous fluorescent light at 22ºC.
3.2 Scanning electron microscopy (SEM)
Plants were fixed in FAA (70% ethanol 89%, formaldehyde 5%, acetic acid 6%) at 4ºC overnight, dehydrated in an ethanol series, critical-point dried in liquid CO2, sputter-coated with gold palladium, analyzed and photographed with a Philips XL 30 FEG SEM.
3.3 Determination of Malonaldehyde (MDA) levels
MDA determination was followed the method described by Zhang (Zhang et al., 2009). Fresh leaves of 20-d-old plants (0.2 g) were homogenized in 6 mL 10% (w/v) trichloroacetic acid. The homogenates were centrifuged at 5000×g for 10 min. A reaction mixture of the supernatant (2 mL) and 2 mL thiobarbituric acid (0.6%) was incubated in a boiling water bath for 15 min, then cooled immediately before centrifugation. Absorbance of the supernatants was determined at 450, 532 and 600 nm, respectively. Calculation of MDA was based on the following formula: C (μ mol /L) = 6.45(A532 - A600) - 0.56 A450.
3.4 Total RNA and genomic DNA extraction
Total RNA was isolated using Trizol reagent (Tiangen, Shanghai) plus 5 mol /L NaCl to reduce polysaccharide level in the RNA. For Arabidopsis RNA isolation, total RNA was extracted from the above ground tissues. And RNA was pretreated with RNase-free DnaseI (Takara, Japan) to remove contaminating genomic DNA. Arabidopsis and narcissus genomic DNA was isolated by using the lysis buffer (0.2 mol /L Tris, 0.05 mol /L EDTA, 7 mol /L Urea, 2% Sarkosyl, pH 8.0 ), employing phenol/chloroform followed by precipitation of DNA with iso-propyl alcohol.
3.5 Isolation and sequence analysis of NTAG1
To obtain the main fragment of the C-lineage genes from both narcissus varieties, primers AGAMOUS F (5'-ctc gag ATG GGG AGG GGT AAG ATA GAG ATC AA-3') and AGAMOUS R (5'-ctc gag TCA TCC CAG TTG AAG GGT AGT-3') were designed according to the NAG gene reported by Wang (Wang et al., 2006). Both the specific 5’ and 3’ primers for NTAG1 contained the generated KpnI recognition site (5- ctc gag -3, a lowercase letter) to facilitate the cloning of this gene. cDNA was synthesized from 500 μg total RNA using a cDNA synthesis kit (TOYOBO, Japan). PCR reactions were performed using the pfu PCR kit (Takara, Japan) and the amplified fragments were purified with the DNA Gel Extraction kit (Tiangen, Shanghai), cloned into the pMD18-T vector (Takara, Japan), and verified by sequencing.
3.6 Plant transformation and transgenic plants analysis
A KpnI fragment containing the full-length cDNA for NTAG1 gene was introduced into the binary T-DNA vector pMon530 (Monsanto, USA), driven by the 35S promoter. The sense orientation construct for this gene was determined using digestion and sequencing. The construct was transferred into Agrobacterium tumefaciens GV3101 (Koncz and Schell, 1986), and then introduced into Arabidopsis plants using the Floral dip method (Clough and Bent, 1998). Transformants that survived in the medium containing kanamycin (50 μg.mL-1) were further verified by PCR and reverse transcriptase-PCR analyses.
The phenotypic effects of NTAG1 in transgenic plants were analyzed in the T1 generation and in the homozygous kanamycin-resistant T3 generation plants through photographing with a digital camera or SEM. To compare the phenotypes of the transgenic and wild type plants, at least 20 transgenic plants from 5 independent lines respectivelly and 20 wild-type plants were grown under the same conditions. Plant height, silique length, branch number, leaf number etc. were recorded.
3.7 Semi-quantitative Reverse transcription-polymerase chain reaction (RT-PCR)
Five microgram of total RNA extracted from different plant tissues was used for generating the first strand of cDNA according to the instructions of the Superscript RT (Toyobo, Japan). PCR amplification was performed with gene specific primers. For SAG12 gene, specific forward (5’-CCAATGACGAATTTCGTTCC-3’) and reverse (5’-TGTCGCAATCAACAAGCTGT-3’) primers were used. For NTAG1 gene, specific AGAMOUS F and AGAMOUS R primers were used. Expression levels of ACTIN were monitored to serve as a quantifying control.
Authors’ contributions
DXJ and XLJ carried out the gene cloning and transgenic planting phenotype analysis, and WY helped with the phenotype analysis and took part in the data analysis. SY took part in the technique instruction. LXF performed the experiment designs and drafted the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This work was supposed by Shanghai Natural Science Program on Key Basic Research Project (09JC1405100) and the National Natural Science Foundation of China (30771155).
References
Bowman J.L., Smyth D.R., and Meyerowitz E.M., 1989, Genes directing flower development in Arabidopsis, Plant Cell, 1(1): 37-52 PMid:2535466 PMCid:159735
Causier B., Castillo R., Zhou J., Ingram R., Xue Y., Schwarz-Sommer Z., and Davies B., 2005, Evolution in action: following function in duplicated floral homeotic genes, Curr Biol., 15(16): 1508-1512 doi:10.1016/j.cub.2005.07.063 PMid:16111944
Coen E.S., and Meyerowitz E.M., 1991, The war of the whorls: genetic interactions controlling flower development, Nature, 353(6339): 31-37 doi:10.1038/353031a0 PMid:1715520
Colombo L., Franken J., Koetje E., van Went J., Dons H.J., Angenent G.C., and van Tunen A.J., 1995, The petunia MADS box gene FBP11 determines ovule identity, Plant Cell, 7(11): 1859-1868 PMid:8535139 PMCid:161044
Colombo L., Franken J., Van der Krol A.R., Wittich P.E., Dons H.J., and Angenent G.C., 1997, Downregulation of ovule-specific MADS box genes from petunia results in maternally controlled defects in seed development, Plant Cell, 9(5): 703-715 PMid:9165748 PMCid:156950
Davies B., Motte P., Keck E., Saedler H., Sommer H., and Schwarz-Sommer Z., 1999, PLENA and FARINELLI: redundancy and regulatory interactions between two Antirrhinum MADS-box factors controlling flower development, Embo J., 18(14): 4023-4034 doi:10.1093/emboj/18.14.4023 PMid:10406807 PMCid:1171478
Drews G.N., Bowman J.L., and Meyerowitz E.M., 1991, Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product, Cell, 65(6): 991-1002 PMid:20174587 PMCid:2823793
Dubois A., Raymond O., Maene M., Baudino S., Langlade N.B., Boltz V., Vergne P., and Bendahmane M., 2010, Tinkering with the C-function: a molecular frame for the selection of double flowers in cultivated roses, PLoS One, 5(2): e9288 doi:10.1371/journal.pone.0009288 PMid:16428599 PMCid:1400554
Ferrario S., Shchennikova A.V., Franken J., Immink R.G., and Angenent G.C., 2006, Control of floral meristem determinacy in petunia by MADS-box transcription factors, Plant Physiol, 140 (3): 890-898 doi:10.1104/pp.105.072660 PMid:8592746
Gan S., and Amasino R., 1995, Inhibition of leaf senescence by autoregulated production of cytokinin, Science, 270: 1986-1988 doi:10.1126/science.270.5244.1986
Gao Z.-m., Chen D.-f., Li X.-p., Cai C.-j., and Peng Z.-h., 2008, Cloning and Sequence Analysis of a Flowering-related M ADS-box Gene in Narcissus tazetta var. chinensis Roem, Acta. Horticuhurae Sinica, 35(2): 295-230 PMid:7919989 PMCid:160514
Jofuku K.D., den Boer B.G., Van Montagu M., and Okamuro J.K., 1994, Control of Arabidopsis flower and seed development by the homeotic gene APETALA2, Plant Cell, 6(9): 1211-1225
Koncz C., and Schell J., 1986, The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel Agrobacterium binary vector, Mol. Gen. Genet, 204: 383-396 doi:10.1007/BF00331014
Lohmann J.U., and Weigel D., 2002, Building beauty: the genetic control of floral patterning, Dev Cell, 2(2): 135-142 doi:10.1016/S1534-5807(02)00122-3 PMid:19033361 PMCid:2615621
Mandel M.A., Bowman J.L., Kempin S.A., Ma H., Meyerowitz E.M., and Yanofsky M.F., 1992, Manipulation of flower structure in transgenic tobacco, Cell, 71(1): 133-143 PMid:9090883 PMCid:156926
Melzer R., Verelst W., and Theissen G., 2009, The class E floral homeotic protein SEPALLATA3 is sufficient to loop DNA in 'floral quartet'-like complexes in vitro, Nucleic Acids Res., 37(1): 144-157 doi:10.1093/nar/gkn900
Mizukami Y., and Ma H., 1992, Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity, Cell, 71(1): 119-131 PMid:9783581
Mizukami Y., and Ma H., 1997, Determination of Arabidopsis floral meristem identity by AGAMOUS, Plant Cell, 9(3): 393-408 PMid:10821278
Murashige T., and Skoog F., 1962, A revised medium for rapid growth and bioassays with tobacco tissue cultures, Physiol Plant 15: 473-479 doi:10.1111/j.1399-3054.1962.tb08052.x
Parcy F., Nilsson O., Busch M.A., Lee I., and Weigel D., 1998, A genetic framework for floral patterning, Nature, 395(6702): 561-566 doi:10.1038/26903
Pelaz S., Ditta G.S., Baumann E., Wisman E., and Yanofsky M.F., 2000, B and C floral organ identity functions require SEPALLATA MADS-box genes, Nature, 405(6783): 200-203 doi:10.1038/35012103 PMid:7549482 PMCid:160949
Pelaz S., Tapia-Lopez R., Alvarez-Buylla E.R., and Yanofsky M.F., 2001, Conversion of leaves into petals in Arabidopsis, Curr Biol., 11(3): 182-184 doi:10.1016/S0960-9822(01)00024-0 PMid:9778845
Roeder A.H., and Yanofsky M.F., 2001, Unraveling the mystery of double flowers, Dev. Cell, 1 (1): 4-6 doi:10.1016/S1534-5807(01)00013-2 PMid:11732606
Rounsley S.D., Ditta G.S., and Yanofsky M.F., 1995, Diverse roles for MADS box genes in Arabidopsis development, Plant Cell, 7(8): 1259-1269
Rutledge R., Regan S., Nicolas O., Fobert P., Cote C., Bosnich W., Kauffeldt C., Sunohara G., Seguin A., and Stewart D., 1998, Characterization of an AGAMOUS homologue from the conifer black spruce (Picea mariana) that produces floral homeotic conversions when expressed in Arabidopsis, Plant J., 15(5): 625-634 doi:10.1046/j.1365-313x.1998.00250.x PMid:12481066 PMCid:166694
Saedler H., Becker A., Winter K.U., Kirchner C., and Theissen G., 2001, MADS-box genes are involved in floral development and evolution, Acta Biochim Pol., 48(2): 351-358 PMid:17794879
Theissen G., 2001, Development of floral organ identity: stories from the MADS house, Curr Opin Plant Biol., 4(1): 75-85 doi:10.1016/S1369-5266(00)00139-4
Tzeng T.Y., Chen H.Y., and Yang C.H., 2002, Ectopic expression of carpel-specific MADS box genes from lily and lisianthus causes similar homeotic conversion of sepal and petal in Arabidopsis, Plant Physiol, 130(4): 1827-1836 doi:10.1104/pp.007948 PMid:1675158
Wang Z.-k., Gao J., Li L.-b., and Peng Z.-h., 2006, Isolation and characterization of the AGAMOUS homologous gene NTAG in Chinese narcissus (Narcissus tazetta var. chinensis Roem), For Stud China, 8(1): 21-26
Weigel D., Alvarez J., Smyth D.R., Yanofsky M.F., and Meyerowitz E.M., 1992, LEAFY controls floral meristem identity in Arabidopsis, Cell, 69(5): 843-859 PMid:1356631
Weigel D., and Meyerowitz E.M., 1993, Activation of floral homeotic genes in Arabidopsis, Science, 261(5129): 1723-1726 doi:10.1126/science.261.5129.1723 PMid:1356630
Zhang Z., Qu W., and Li X., 2009, Instructions and techniques of plant physiological experiment (in Chinese), Higher Education Press, Beijing, China, pp.227-229 PMid:1350515
. PDF(657KB)
. FPDF
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Xinjie Deng
. Lijun Xiong
. Yang Wang
. Xiaofang Li
Related articles
. Chinese narcissus AGAMOUS homologue
. NTAG1
. Flower development
. Senescence
Tools
. Email to a friend
. Post a comment