Genetic Diversity of 30 Cai-xins (Brassica rapa var. parachinensis) Evaluated Based on AFLP Molecular Data 
* The authors who contribute equally
 Author
Author  Correspondence author
Correspondence author
Molecular Plant Breeding, 2011, Vol. 2, No. 7 doi: 10.5376/mpb.2011.02.0007
Received: 18 Feb., 2011 Accepted: 12 Apr., 2011 Published: 12 Apr., 2011
Shi et al., 2011, Genetic Diversity of 30 Cai-xins (Brassica rapa var. parachinensis) Evaluated Based on AFLP Molecular Data, Molecular Plant Breeding Vol.2 No.7 (doi: 10.5376/mpb.2011.02.0007)
Cai-xin is common Chinese name for Brassica rapa var. parachinensis that is one of very important leafage vegetables in South China. The objectives of this research were to detect genetic diversities of the selected 30 Cai-xins based on AFLP makers as well as to evaluate the feasibilities of AFLP approach for biodiverse study . In this paper, the 25 pairs of AFLP primers were employed to generate 1160 amplified bands, of which 876 bands account for 76% were polymorphic bands The average polymorphism of tested 25 primer combinations reached 80%, the mean of polymorphism information content (PIC) was about 0.0239 and the amount of polymorphic loci was about 85.33%. The parameters of genetic diversity were calculated by the aid of GenAlEx 6.4 software including the number of different alleles (Na) 1.754, the number of effective alleles (Ne) 1.544, Shannon's Information Index (I) 0.472 and He 0.363. The values of genetic distance (GD) and genetic similarity (GS) were 0.112 and 0.895, respectively. Statistic analysis revealed that almost 100 percentage of variation existed within Cai-xins based on AMOVA data. The tested Caixins can classified into four groups at the 0.17 threshold of Nei’s genetic distances by clustering analysis based on UPGMA approach . The present results indicated that the genetic diversity of the tested Cai-xins should be quite low and the genetic variations of Caixins be mostly attributed to within varieties. Whereas we confidentially have conclusions that AFLP approach might be useful, efficiency and accuracy to detect genetic diversity among varieties, landraces and lines of Caixin, especially for those which have close relationship.
Molecular markers have been widely applied to detect genetic diversity in Brassica vegetables due to the polymorphisms of the genomic DNA at the molecular marker level. There were some studies on the genetic diversity of Caixin detected by the molecular makers of AFLP, RAPD, SRAP and ISSR (Chu., 2002; Guo et al., 2002; Zhao et al., 2005; Han, 2008; Ma, 2008; Tan et al., 2009; Sun et al., 2010). As known, RAPD, randomly ampliï¬ed polymorphic DNA, is generated by single, short and arbitrary oligonucleotide primer in length of 8~10 bp, and ISSR, Inter-simple sequence repeat polymorphism is generated with single primer which is recognized as an abundant simple sequences existing in genome (Tautz et al., 1984). Whereas AFLP, as well-known molecular marker, might uncover much more DNA polymorphism because of the limitation of the restriction endonuclease-digestion procedures although the procedures of polyacrylamide gel running and silver stain are significantly elevate the band polymorphism and segregating precision (Blears et al., 1998). AFLP marker is a dominant molecular marker and only needs anonymous genomic DNA for PCR performance, the PCR results has much more comparable with whose mentioned above at a certain extent (Zhang and Yang, 2008). So far AFLP has been widely used to detect genetic diversities of Brasscia vegetables (Sun et al., 2001; Guo et al., 2002; Chu, 2002; Zhao et al., 2005).
Cai-xin is common Chinese name for Brassica rapa var. parachinensis that is one of very important leafage vegetables in South China. Caixin has been characterized based on the morphological diversities with regard to agronomic traits including leaves, stems and inflorescences as well as the edible parts. The traits of the colors, size of stem and flowering time are known as important morphological characters, which were recognized as the criteria for classification and identification of commercial products. The classification of Caixin based on morphological approach has somewhat differences with that of Brassica vegetable based on molecular marker (Guo et al., 2002; Yang et al., 2006; Wang, 2006; Sun et al., 2010). The reasons might refer to close relatives between species. The previous studies RAPD and ISSR motioned above were prevalent for the classification of Caixin, which usually assigned Caxin to the independent group or group together with other non-heading Chinese cabbage. The reason for this might be caused by the long term artificial selections, plant introduction or natural variation (He et al., 2002; Wang, 2006; Sun et al., 2001; Guo et al., 2002; Chu, 2002). It is obvious that genomic differences between Caixin and other Brassica vegetables need to be identified. As we know that artificial selection is based on the natural and heritable variation in systematic breeding program, which is dominant approach in the Caixin breeding (Ma and Wang, 2006), how to distinguish the variations or genetic diversities among Caixin germplasms might be still a big challenge to the breeders while they try to explore and widen their genetic diversities in the future.
The previous results came from RAPD and ISSR indicated Caixin’s genetic diversity was comparatively low and there was no obvious grouping pattern in classification at molecular level (Qiao et al., 1999; He et al., 2002; Wang, 2006; Han et al., 2008; Ma, 2008; Tan et al., 2009; Zhang et al., 2010; Sun et al., 2010). There is few information about Caixin’s genetic diversity based on AFLP data (Chu, 2002; Zhao et al., 2005). In present research we selected 30 Caixin collections from different sources to detect Caixin genetic diversities using AFLP approach and to evaluate the feasibilities of AFLP approach for biodiverse study.
1 Results
1.1 AFLP polymorphism
In present study, 30 Caixins were analyzed with 25 selective primer combinations of AFLP (Table 1). Total 116 0 bands were amplified in which 876 bands were polymorphic bands, accounting for 76%. The sizes of amplified bands ranged from 100 bp to 1 500 bp, while most of specific bands are located in the ranges from 700 bp to 1 000 bp. The average number of polymorphic bands for each primer combination is 44 in the range from 13 to 84. The mean polymorphism rate of each primer combination was up to 80% in the range from 56.8% to 97.1%. The PIC values for each primer combination varied from 0.011 6 to 0.031 7 that the mean was 0.023 9. These parameters showed that there were sufficient AFLP polymorphisms existing in selected Caixin germplasms. Each primer combination used in this research could detect the genetic diversity of Caixin because there were significant differences existing between the tested primer combinations. To be as case, primer combination, E-ATG/M-CTG, could reveal 97% AFLP polymorphism in selected tested Caixin germplasms.
.png) Table 1 Amplification results of 25 AFLP primer combinations in 30 Caixins |
1.2 Genetic diversity analysis
GenAlEx 6.4 software was employed to calculate 30 Caixin’s genetic diversity (Table 2). The average percentage of polymorphic loci was 85.33% in the range from 51.6% (Lvbao 701) to 91.78% (Guiliu October). The following parameters are including as Na, Ne, I, He, GD and Gs. That is, the number of different alleles (Na) 1.754 from 1.032 (lvbao 701) to 1.904 (Guiliu October), the number of effective alleles (Ne) 1.544 from 1.211 (lvbao 701) to 1.665, Shannon’s Information Index (I) 0.472 from 0.232 (lvbao 701) to 0.543 (Guiliu October) and He 0.363 from 0.165 (lvbao 701) to 0.426. The values of genetic distance (GD) and genetic similarity (GS) were 0.112 from 0.026 (CX 9-1 and CX 6-14) to 0.275 (Lvbao 701 and Australia 008) and 0.895 from 0.76 (Australia 008 and Caixingli Si Jiu) to 0.959, respectively.
 Table 2 The parameters of genetic diversity of 30 caixins based on AFLP data |
Analysis of Molecular Variance (AMOVA) for the 30 Caixins based on AFLP data derived from 876 markers indicated that almost 100 percentage of variation was contributed by Caixin varieties (Φpt=0.006, P>=0.227) and lines (Φpt=0.005, P>=0.653), respectively.
1.3 Cluster analysis
The dendrogram for 30 Caixins was constructed according to unweighted pair group method of arithmetic averages (UPGMA) and Nei’s genetic distance. The four groups were classified at the 0.17 threshold of Nei’s genetic distances by clustering analysis based on UPGMA approach (Figure 1). Group I was composed of eleven germplams, CX6-6, CX6-6-1, CX6-7, CX6-13, Si Jiu-19, CX5-12, CX5-14, CX9-1, CX9-4, Youqing no. 12, Lvbao 701. Group II was composed of 15 germplasms, CX5-3, CX5-4, Dong Guan 45 Days, Australia 50 Days, Kuaida 28 Days, CX3-6, New Zealand Si Jiu, Caixingli Si Jiu, CX3-6-1, Australia 608, Australia 008, Hong Kong 45 Days, Mingyou 308 sweet, No. 31, GX3-1. Group III was composed of Youxin no. 2, GX6-14 and Guiliu October. And Group IV was Teqing 60 Days only. The results showed that most lines were grouped together with at least one of their parents. Such as the serials lines of CX6 were clustered with Si Jiu-19. New Zealand Si Jiu, Si Jiu-19, Caixingli Si Jiu grouped in one cluster. The results indicated there is very close relationship in the tested Caixin germplasms.
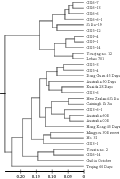 Figure 1 The dendrogram of the selected 30 Caixins based on UPGMA |
2 Discussion
To assess the genetic diversity and genetic relationship is essential for effectively utilizing genetic germplasms in the breeding program. The availability of agronomic morphological traits in Caixin are so limited that molecular markers should genuinely play more important roles in breeding program. Most of studies on Caixin varieties and even mutant lines derived from the induction by carrying Space Shuttle were conducted by RAPD or and ISSR (Ma, 2008). Although RAPD, ISSR and AFLP have the common vantages that they do not need to any background information of the genome in advance, the results of the amplication are quit different even based on same genomic DNA. In present work, AFLP generated 1 160 bands by 25 primer combinations. 876 bands were polymorphic. It is obvious that AFLP could reveal much more polymorphisms and that is more effective (Wang, 2006; Tan et al., 2009; Zhang et al., 2010; Sun et al., 2010). The PIC values indicated that the efficiency of primer combinations was very high and most of primer combinations had relatively high discriminatory ability. Due to the significant correlationship and comparability between RAPD and AFLP (Zhang and Yang, 2008), comparative studies on the genetic variation had been conducted among AFLP, RAPD (Han et al., 2008; Ma, 2008; Tan et al., 2009), ISSR (Ma, 2008; Sun et al., 2010,) and SRAP (Ma, 2008). The results showed that the parameters, such as Na, Ne, He, I calculated based on AFLP data were higher than that of others. This results illustrated that AFLP had more powerful to uncover more genetic diversity. AMOVA of AFLP in this work revealed almost 100% genetic variation existing within whatever varieties or lines, which suggested that strong selection pressures might have contributions to the low levels of genetic diversity in Caixin. Be a kind of non-heading variety, Xiaobaicai, Brassica chinensis, the results of AMOVA was coincidence with another research results done by SSR in which variation was significant contribution by inter-variety variance too (Yim et al., 2009). In other words, factors such as geography may have little effect on genetic variation within varieties. The reason may be raised that systematic breeding is a conventional and major approaches for the development of new Caixin variety, while recurrent selection within Caixin is inevitable to bring about close genetic relationship and high homogenizations in breeding program. Anyway, the conclusion of low genetic diversity and close genetic relationship within Caixin by AFLP analysis was consistent with that by RAPD and ISSR (Tan et al., 2009; Sun et al., 2010).
GD and GS are important indexes to evaluate the relationship among varieties. The former indicates the difference between varieties, the greater of the GD the more difference existing between varieties. Whereas the later is reverse, the greater of the GS the more similar existing between varieties. In our work GD ranged between 0.026 and 0.275 while GS ranged between 0.76 and 0.943. Compared to results based on RAPD (Tan et al., 2009) and ISSR (Sun et al., 2010), as if there were more close relationship between Caixin varieties and lines in this study. AFLP indeed detect this kind of relationship. Cluster analysis divided the 30 Caixins into four groups but cluster pattern was still not very clear. It was same with the researches mentioned above that Caixin varieties often clustered together and isolated from other Brassica vegetables (Guo et al., 2002; Han et al., 2008; Ma, 2008; Tan et al., 2009; Sun et al., 2010). Although we could find something interesting, for example, New Zealand Si Jiu, Si Jiu-19 and Caixingli Si Jiu grouped together, the same way with Australia 608, Australia 008 dark green and Australia 50 Days. These results testified that there must have very close relationship between these varieties. The cluster results also showed that the lines derived from some or other parents grouped together. For instance, CX6-6, CX6-6-1, CX6-7 and CX6-13 grouped together in the cluster I. These results implied that these varieties or lines should have somewhat changes in genome after long term artificial selection. Some lines such as CX3-1, CX3-6 were not classified together, which might imply that these germplasms have distinctly genetic diversity.
In conclusion, AFLP is extremely useful and efficiency and accuracy tool to assess genetic diversity of Caixin varieties or lines, especially for lines which have close relationship although it is considered that the approach is high costs and time consumed and complicated protocol. Compared with previous AFLP analysis, our research work also revealed more polymorphisms not only Caixin’s varieties but also lines used in this study, therefore there were much more information to be provided. In addition, the genetic diversity of Caixin is very low and genetic variations most attribute to varieties. It is without question that molecular markers can enhance the selection precision and breeding efficiency, but to widen genetic fundament may need to be dependent on new germplasm introduction. Therefore we strong believe that novel breeding technologies such as distant hybridization and transgenic technology should be introduced into the Caixin breeding program in order to solve the problem of narrow genetic background.
3 Material and Methods
3.1 Plant materials
Thirty Caixins were employed in this study. These varieties were collected from South China (Guangdong province: Dong Guan 45 Days, Teqing 60 Days, Youqing no. 12, Si Jiu-19, Mingyou 308 sweet, No. 31, Kuaida 28 Days, Youxin no. 2, Lv bao 701; and Guangxi province: Guiliu October, GX3-1, GX3-6, GX3-6-1, GX5-3, GX5-4, GX5-12,GX5-14, GX6-6, GX6-6-1, GX6-7, GX6-13, GX6-14, GX9-1, GX9-4), Hong Kong (Hong Kong 45 Days, Caixingli Si Jiu), Australia (Australia 008, Australia 608, Australia 50 Days) and New Zealand (New Zealand Si Jiu).
3.2 DNA extraction
30 Caixin seeds were planted in the greenhouse. Leaves of young seedling were randomly collected from four individual plants each variety. Genomic DNA was extracted from these mixed leaves by modified CTAB method as follows. One gram of ground fresh leave tissue was suspended in a 1.5 ml centrifugal tube with preheated 750 µl CTAB. The suspension was shaken up, kept at 65℃ for 45 min, and then centrifuged with 12 000 rpm for 10 min. The supernatant was abandoned later. Extracted by chloroform isoamylalcohol (24:1) twice and precipitation with two-thirds volume of isopropanol under -4℃, and then centrifuged at 12 000 rpm for 10 min. The pellet was washed with 70% ethanol twice. Then the pellets were air dried. DNA was dissolved in 50 µl deionized water. The DNA quality was inspected by 1% agarose gel electrophoresis and stored at -20℃.
3.3 AFLP analysis
AFLP was performed on the basis of the protocol introduced by Vos et al. (1995) with a minor modifications (Shi, 2006). Double digested the genomic DNA with EcoR I/MseI. The enzymed DNA was ligated to the adaptors which had the EcoR I/MseI restriction sites at both sides. The preselective amplification was performed with non-selective EcoR I/MseI primer combination. The PCR products were diluted 10-fold with TE and used to be template for selective amplification. The selective amplification primer combination was EcoR+3/Msel+3 (Table 2). The selective amplification PCR products were added with the same volume of loading buffer. Set in 95℃ for 3 min for denature. Put on the ice immediately. Draw 5.5 µl to% (w/v) polyacrylamide gel containing which was preheated for 45 min at 55W. The gels were silver stained (Lu et al., 2001) and placed in the air to dry.
3.4 Data analysis
All distinct bands on AFLP gel were identified and wrote down according to their position independently for each cultivar and primer combination. A matrix was constructed with all these data. The polymorphic rate (P) and Polymorphism information content (PIC) values were calculated as described by Botstein et al (1980). GenAlEx 6.4 software (Rod Peakall. http://www.anu.edu.au/BoZo/GenAlEx/new_version.php) was used to calculate the amount of index of genetic diversity including GS (Genetic similarity), GD (Genetic distance) and AMOVA. Cluster analysis was conducted based on the UPGMA (Unweighted Pair Group Mean Average) method with MEGA4 (http://www.megasoftware.net/).
Authors’ contributions
Weidong Shi fulfilled major part of this paper, including varieties collection and lines selection and cultivation, experiment design and accomplishment, data analysis and manuscript preparation. Ruikui Huang participated in germplasm collection. Shengmao Zhou participated in germplasm collection and experiment design. Faqian Xiong gave great assistance to experimental accomplishment. All authors read and approved the final manuscript.
Acknowledgements
This study was jointly supported by Agricultural Science Institute of Guangxi Basic Research Project (200805Z) and (0815011-6-1) and Natural Science Foundation of Guangxi Zhuang Autonomous Region (2010GXNSFA013084). Mention of trade names or commercial products in this paper is solely for the purpose of providing specific information and does not imply recommendation or endorsement by authors or institutes or university involved in this study.
References
Blears M.J., de Grandis S.A., Lee H., and Trevors J.T., 1998, Amplified fragment length polymorphism (AFLP): a review of the procedure and its applications, J. Indu. Microb. & Biotec., 21: 99-114
Botstein D., White R.L., Skolmick M., and Davis R.W., 1980, Construction of a genetic linkage map in man using restriction fragment length polymorphism, Amer. J. of Hum. Genet., 32: 314-332
Chu L.T., 2002, Studies on genetic polymorphisms with AFLP and taxonomy in Brassica campestris (2n=20), thesis for M.S., Zhejiang University, Supervisor: Cao J.S.
Guo J.X., Zhou N.Y., Ma R., and Cao M.Q., 2002, Genetic diversity in Brassica rapa revealed by AFLP molecular mark, Journal of Agricultural Biotechnology, 10(2): 138-143
Han J.M., Hou X.L., Xu H.M., and Wang J.J., 2008, Rapd analysis of genetic diversity of non-heading chinese cabbage (Brassica campestris. ssp. chinensis makino) germplasm, Journal of Nanjing Agricultural University, 31(3): 31-36
He Y.T., Tu J.X., Fu T.D., Li D.Y., and Chen B.Y., 2002, Genetic diversity of germplasm resources of Brassica campestris. L in china by RAPD markers, Acta Agronomica Sinica, (28): 697-703
Lu G.Y., Yang G.S., and Fu T.D., 2001, Silver-Stained AFLP Novel Assay for DNA Fingerprinting in Brassica napus, Jourtud of Huazhong Agricultural University, 20(5): 413-415
Ma J.J., 2008, Preliminary study of germplasm genetic diversity in non-heading chinese cabbage with molecular marker, Thesis for M.S., Yangzhou University, Supervisor: Bo T.Y., and Chen X.H.
Ma S.M., and Wang Y.F., 2006, Progress on breeding of flowering Chinese cabbage, Northern Horticlture, (3): 40-41
Shi W.D., 2006, Analysis of oilseed rape flowering time variation and cloning of Arabidopsis late flowering time gene, Dissertation for Ph.D., Chinese Academy of Agriculture Sciences, Supervisor: Liu S.Y.
Sun D.L., Zhao Q.C., Song W.Q., and Chen R.Y., 2001, Relationships analysis of Chinese cabbage species by AFLP, Acta Horticulture Sinica, 28(4): 331-335
Sun X.M., Qiao A.M., Sun M., Gui T.Q., and Yin C.X., 2010, ISSR anylysis of genetic diversity of 27 flowering Chinese cabbage, Journal of Southerwest China Normal University (natural science edition), 35(1): 119-123
Tan X., Zhang J.D., Qiao A.M., Li L.F., and Zhen S.W., 2009, RAPD analysis of genetic diversity of flowering Chinese cabbage, Guangdong Agricultural Sciences, 10: 154-157
Vos P., Hogers R., Bleeker M., Reijans M., van de Lee T., Hornes M., Frijters A., Pot J., Peleman J., and Kuiper M., 1995, AFLP: a new technique for DNA fingerprinting, Nucleic Acid Research, 21: 4407-4414 doi:10.1093/nar/23.21.4407
Wang L., 2006, Studies on genetic diversity of flowering Chinese cabbage (Brassica campestris L. ssp. chinensis var. utilis tsen et lee) by RAPD analysis, Thesis for M.S., Southwest University
Yang H., Liu Z.C., Chen H.R., Chen L., and Luo L.J., 2006, Genetic diversity analysis based on SSR markers of Brassica vegetable germplasm distribution in ShangHai, Journal of Plant genetic resources, 7(3): 264-269
Yim G.R., Ding D.R., Wang C.P., Leong W.H., and Hong Y., 2009, Microsatellite markers to complement distinctness, uniformity, stability testing of Brassica chinensis (Xiao Baicai) varieties, The Open Horticulture Journal, 2: 54-61 doi:10.2174/1874840600902010054
Zhang D.Q., and Yang Y.P., 2008, A statistical and comparative analysis of genetic diversity detected by different molecular markers, Acta Botanica Yunanica, 30(2): 159-167
Zhang J.Y., Zhang J.D., Huang H.D., Zhang H., Yin C.X., Qiao A.M., and Zheng Y.S., 2010, RAPD analysis for space-induced mutant lines of Brassica parachinensis bailey, Guangdong Agricultural Sciences, 1: 35-42
Zhao J.J., Wang X.W., Deng B., Lou P., Wu L., Sun R.F., Xu Z.Y., Vromans J., Koornneef M., and Bonnema G., 2005, Genetic relationships within Brassica rapa as inferred from AFLP ï¬ngerprints, Theoretical and Applied Genetics, 110: 1301-1314 doi:10.1007/s00122-005-1967-y
. PDF(201KB)
. FPDF
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Weidong Shi
. Ruikui Huang
. Shengmao Zhou
. Faqian Xiong
Related articles
. Caixin ( Brassica rapa var. parachinensis )
. AFLP
. Genetic diversity
. Polymorphism
Tools
. Email to a friend
. Post a comment


