2. Plant Breeding and Biotechnology Division, Philrice, Science City of Munoz, Nueva Ecija, 3119, Philippines
 Author
Author  Correspondence author
Correspondence author
Molecular Plant Breeding, 2013, Vol. 4, No. 38 doi: 10.5376/mpb.2013.04.0038
Received: 03 Sep., 2013 Accepted: 22 Nov., 2013 Published: 30 Dec., 2013
Estimated of linkage disequilibrium (LD) are important as an indication of how useful LD-base association genetics approaches can be when compared with other available mapping methods. The success of association mapping efforts depends on the possibilities of separating LD due to linkage from LD due to other causes. In an attempt to associate markers with drought tolerance at vegetative stage, we examined the pattern of LD in a diversity and stress adaptation rice panel containing 184 rice germplasm accessions with 141 polymorphic SSR markers that were nearly evenly distributed at 3 mb bin on the 12 rice chromosomes. Significant LD was detected across the genome of the 184 rice genotypes and the extent of LD varied with different chromosomes, selfing in O. sativa species and existing of population structure in the rice panel might be the major factors of creating high LD. No genetic linkage was detected except for two pairs of SSR markers RM288 and RM464 (11.98758 Mb), RM215 and RM464 (14.61396 Mb), which were located on chromosome 9 at a relatively far distance but showed significantly higher correlation. The high degree of LD and fast decay of LD detected in this experiment indicating the 141 SSR markers used in the experiment could be feasible to carry out the whole-genome scan association studies with a relatively high resolution.
In the last decades or so, the conventional genetic based strategies have been used successfully in QTL (quantitative trait loci) mapping for agronomically and economically important genes in different crops species such as tomato, Arabidopsis and rice, however, QTLs identified throught this method is limited to loci with large effects on the target quantitative trait variation, other techniques that enable the rapidly identification of genes playing modest roles associate with the variation of quantitative traits are also needed. Association mapping via linkage disequilibrium or LD (non-random association of alleles at different loci) offers promise in this area. The traditional approach of linkage/QTL mapping reliant on developing large mapping populations continues to suffer from lack of mapping resolution inherent in samples with limited meiotic cross-over events, while in association mapping, there may not be any need to make crosses initially to generate segregating populations, the natural variation that exists in the available germplasm can be utilized for mapping straightaway (Oraguzie et al., 2007).
LD plays a central role in association analysis, estimated of LD are important as an indication of how useful LD-base association genetics approaches can be when compared with other available mapping methods (Rafalski et al., 2004), the distance over which LD persists will determine the number and density of markers and experimental design needed to perform an association analysis. Garris et al. (2003) found out rapid decay of LD around the Xa5 locus in rice (Oryza sativa L.) happed at each 100 kb as a result of more recombination events, under this condition an average of one marker per centiMorgan (where
In an attempt towards association mapping for drought tolerance at vegetative stage in a diversity and stress adaptation rice panel, we had already examined the phenotypic performance of 184 rice germplasm accession for vegetative drought tolerance under natural drought conditions (Xiao et al., 2012a) and its population structure and genetic relatedness using 141 polymorphic SSR markers that were nearly evenly distributed at 3mb bin on the 12 rice chromosomes (Xiao et al., 2012b), with the same genotypic data, we were now trying to find out the LD model in this rice panel and to check out if a whole-genome-scanning association mapping could be used to identify loci for drought tolerance at vegetative stage.
1 Results
.png)
Figure 1 Bands produced by SSR markers. A: RM192; B: RM112; C: RM595
Significant LD was detected across the genome of the 184 rice genotypes and extent of LD varied with different chromosome (Figure 2). The pair-wise r2 among the 141 SSR markers varied from 0.0000 to 0.78197, the 95th percentile of the distribution of these estimates was 0.3886, and it was used as a population-specific threshold for this parameter as an evidence of linkage, value of r2>0.3886 were probably due to genetic linkage.
.gif)
Figure 2 LD between 141 SSR markers
No genetic linkage was detected among the 141 SSR markers across the 12 chromosome, however, two pairs of SSR markers RM288 and RM464 (11.98758 Mb), RM215 and RM464 (14.61396 Mb) showed significantly higher correlation at far distance (Figure 3), all the three SSR makers were located on chromosome 9.
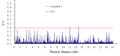
Figure 3 Linkage disequilibrium (r2) as a function of physical distance (Mb) among 141 markers
2 Discussion
In cultivated barley genome-wide LD extended from
Mather et al (2007) also reported that the extent of linkage disequilibrium differs in significant way between domesticated Asian rice and O. rufipogon, both in genome wide LD and in targeted genomic regions, higher LD was found in O. sativa variety groups. In the presence of high LD, lower marker density is required for a target region with greater potential for detecting markers strongly associated with the target gene polymorphism, even if distant physically (Agrama et al., 2007), whole-genome-scan association study is feasible in this case.
The success of association mapping efforts depends on the possibilities of separating LD due to linkage from LD due to other causes. Factors such as the mating system, the recombination rate, population structure, population history, genetic drift, directional selection, and gene fixation at different rates on different chromosome regions can all affect the patterns of LD (Gaut and Long, 2003). In general, LD decay with distance occurs at a much slower rate in self pollinated plants, such as Arabidopsis, rice, barley, durum wheat, and sorghum, than in outcrossing species. In this study, LD decayed faster than 3 MB bin, Rakshit et al. (2007) reported an LD decay of ~50Kb in indica and of ~5 Kb in O.rufipogon, which was significantly smaller as compared with 3 MB in this study, a fast decay in the genome, however, is an inference of high resolution for association analysis.
However, two pairs of markers RM288 and RM464 (11.98758 Mb), RM215 and RM464 (14.61396 Mb) all were located on chromosome 9 but at relatively far distance showed significant linkage. The same phenomenon also reported by Skot et al. (2005) in the natural population of perennial ryegrass at a genome-wide scale using AFLP genetic markers in which the majority of the linked pairs were in significant LD within genetic distance of
3 Conclusion
4 Materials and Methods
.png)
Table 1 SSR markers at each 3 Mb bin used in the experiment
LD was analyzed followed the way used by Flavio and Mark (Flavio and Mark, 2006), in detail: the program TASSEL (http://www.maizegenetics.net) was used to estimate the LD parameter r2 among loci and the comparison-wise significance was computed by 1000 permutations. The unlinked r2 and the syntenic r2 were estimated separately based on the LD for unlinked loci and for loci on the same chromosome. A critical value of r2 derived from the distribution of the unlinked r2 by square root transformed the unlinked estimates was used as an evidence of linkage. The parametric 95th percentile of the distribution was taken as the population-specific critical value of r2, beyond which LD was considered to be caused by genetic linkage and the interception of the syntenic r2 with this baseline was considered as the estimate of the extent of LD in the chromosome.
Acknowledgments
References
http://dx.doi.org/10.1007/s11032-006-9066-6
Flavio B., and Mark E.S., 2006, Association Mapping of Kernel Size and Milling Quality in Wheat (Triticum aestivum L.) Cultivars, Genetics, 172: 1165-1177.
Garris A.J., McCouch S.R., and Kresovich S., 2003, Population Structure and its Effects on Haplotype Diversity and Linkage Disequilibrium Surrounding the Xa5 locus of Rice (Oryza sativa L.), Genetics, 165: 759-769.
Gaut B.S., and Long A.D., 2003, The lowdown on linkage disequilibrium, Plant Cell, 15: 1502-1506.
http://dx.doi.org/10.1105/tpc.150730
Liu K.J., and Muse S.V., 2005, Power Marker: An Integrated Analysis Environment for Genetic Marker Analysis, Bioinformatics, 21: 2128-2129
Mather K.A., Caicedo A.L, Polato N.R., Olsen K.M., McCouch S., and Purugganan M.D., 2007, The extent of linkage disequilibrium in rice (Oryza sativa L.), Genetics, 177(4): 2223-2232.
http://dx.doi.org/10.1534/genetics.107.079616
Murray M.G., and Thompson W.F., Rapid isolation of high molecular weight plant DNA, Nucleic Acid Res., 1980, 8: 4321-4325
Oraguzie N.C., Rikkerink E.H.A., and Gardiner S.E., 2007, Nihal De Silva H. Association Mapping in Plants, Springer Science + Business Media, LLC
Oraguzie N.C., Phillip L.W., Rikkerink E.H.A., and Silva H.N.D., 2007, Linkage Disequilibrium: in Association Mapping in Plants. Springer Science + Business Media, LLC
Oraguzie N.C., and Phillip L.W., 2007, An Overview of Association Mapping: in Association Mapping in Plants. Springer Science + Business Media, LLC
Palaisk K., Morgante M., Williams M., and Rafalski A., 2003, Contrasting Effects of Selection on Sequence Diversity and Linkage Disequilibrium at two Phtoene Synthease Loci, Plant Cell, 15:1795-1806
Rafalski A., and Morgante M., 2004, Corn and humans: recombination and linkage disequilibrium in two genomes of similar size, Trends Genet., 20(2): 103-111
Rakshit S., Rakshit A., Matsumura H., Takahashi Y., and Hasegawa Y., 2007, Large-Scale DNA Polymorphism Study of Oryza sativa and O. rufipogon Reveals the Origin and Divergence of Asian Rice, Theor Appl Genet., 114:731-743
Skot L., Humphreys Mo, Armstead I., Heywood S., Skot K.P., Sanderson R., Thomas I.D., Sanderson R., CHorlton K.H., and Hamilton N.R.S., 2005, An association mapping approach to identify lowering time genes in natural populations of Lolium perenne (L.), Mol. Breed., 15:233-245
http://dx.doi.org/10.1007/s11032-004-4824-9
Xiao Y.L., Lei J.G., Yu C.Y., Quirino D.D.C., Jonalyn M.C., and Dindo A.T., 2012, Genetic Similarity and Population Structure in a Rice Drought Stress Adaptation Panel, Acta Agricultural Universitatis Jiangxiensis, 34(5): 886-892
Xiao Y.L., Yu C.Y., Lei J.G., Quirino D.D.C., Jonalyn M.C., and Dindo A.T., 2012, Screening of Rice Germplasm Accessions for Vegetative Drought Tolerance. Acta Agricultural Universitatis Jiangxiensis, 34(3): 0428-04
. PDF(712KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Yulong Xiao
. Chuanyuan Yu
. Jonalyn M. Yabes
. Jianguo Lei
. Xiaoling Wang
. Zhiquan Wang
. Hongping Chen
. Dindo A. Tabanao
Related articles
. Linkage disequilibrium (LD)
. LD decay
. Rice
. Association mapping
Tools
. Email to a friend
. Post a comment


