2 Bioscience and Bioengineering School, Hebei University of Economics and Business, Shijiazhuang, 050061
3 Graduate University of the Chinese Academy of Sciences, Beijing, 100049
 Author
Author  Correspondence author
Correspondence author
Molecular Plant Breeding, 2011, Vol. 2, No. 9 doi: 10.5376/mpb.2011.02.0009
Received: 16 Mar., 2011 Accepted: 03 May, 2011 Published: 21 May, 2011
Pan et al., 2011, Evolution of the Genes Encoding Starch Synthase in Sorghum and Common Wheat, Molecular Plant Breeding Vol.2 No.9 (doi: 10.5376/mpb.2011.02.0009)
Starch synthases (SSs) play important roles in plant starch synthesis. Five enzymetic isoforms of starch synthases have been found in plant, some of isoforms have further diverged possibly via whole genome duplication events, which might result in two or three subclasses of the sequences in rice and maize. In this study, we found the retention of GBSS, SSII, and SSIII duplicators except for the SSIV duplicator in genomes of sorghum and wheat. The SSIV gene might have been lost in maize and sorghum genomes based on the synteny relationship among rice maize and sorghum. Expression analyses indicated that the SS duplicators were also diverged from the duplicators of SbGBSSI, SbSSIIIa in expression pattern, and SbSSIIa were expressed mainly in endosperm in sorghum, whereas SbGBSSII, SbSSIIb, and SbSSIIIb of sorghum, TaSSIIb and TaSSIIIb of common wheat, were expressed mainly in leaves. Our findings, in combination with previous studies, indicate that the SSIV duplicator should be not remained in all Gramineae species, while the expression of duplicated SS genes diverged similarly in the studied species.
Whole genome duplication (WGD) or polyploidy event is a prominent process in Gramineae plants and has been signiï¬cant in the evolution and separation history (Wendel, 2000). Interdisciplinary approaches combining phylogenetic and structural genomic data suggest that the Gramineae genes have undergo a WGD about 70 million years ago (Mya) before the divergence of the Gramineae (Paterson et al., 2004). In addition, it also found that the maize genome is the product of a genomic allotetraploid event approximately 12 Mya ago (Gaut and Doebley, 1997). Subsequently, the maize genome ‘diploidized’ by deleting most of the duplicated centromere regions and deleting or tolerating the degeneration of one number of most of its paired genes (Song and Messing, 2003; Brunner et al., 2005).
Starch is the main storage carbohydrate in plants and also by far the major carbon source in Gramineae seeds and is used as a primary store of energy for metabolism and biosynthesis. Starch is composed of glucose (Glc) polymer that occurs in two main forms: amylose and amylopectin. Both type starches are synthesized inside plastids in higher plants, and are achieved through the coordinated interactions of several of starch biosynthetic enzymes, including ADP glucose pyrophosphorylase (AGPase), starch synthase (SS), starch branching enzyme (BE), and starch debranching enzyme (DBE) (Ball and Morell, 2003). The duplicate sets of some genes which involved in the core pathway of starch biosynthesis were retained in rice, maize, and wheat following the ancient WGD event in Gramineae (Harn et al., 1998; Vrinten and Nakamura, 2000; Hirose and Terao, 2004; Dian et al., 2005).
The starch synthase (SS, EC 2.4.1.21) catalyzes the synthesis of the glucan polymers by transfering the glucosyl moiety from ADP-Glc to the nonreducing end of a preexisting α-1,4 linked glucan primer. Multiple isoforms of SSs are found in plants (Smith et al., 1997; Ball et al., 1998; Hirose and Terao, 2004). The granule-bound starch synthase family (GBSS) is responsible for amylose synthesis and is exclusively bound to the starch granule. The SSâ… -â…¢ isoforms are involved in amylopectin biosynthesis, while the SSâ…£ isoform in the control of granule numbers (Ball and Morell, 2003; Roldán et al., 2007).
SS isoforms have been identiï¬ed in several Gramineae genomes through amino acid homology analysis. In rice (Oryza sativa L.), there are 10 SS isoforms separated into ï¬ve types, including two GBSS genes (GBSSâ… /WX and GBSSâ…¡), one SSâ… gene, three SSâ…¡ genes (SSâ…¡a, SSâ…¡b, and SSâ…¡c), two SSâ…¢ genes (SSâ…¢a and SSâ…¢b), and two SSâ…£genes (SSâ…£a and SSâ…£b) (Hirose and Terao, 2004). Compared to the rice genome through the current data, Maize (Zea mays L.) genome contains two SSâ…¡b and two SSâ…¢b genes, but only one SSâ…£ gene (Yan et al., 2009). In wheat, seven SS genes, GBSSâ… /WX, GBSSâ…¡, SSâ… , SSâ…¡a, SSâ…¡c, SSâ…¢a, and SSâ…£b, had been cloned (Clark et al., 1991; Li et al., 1999a, 1999b, 2000; Leterrier et al., 2008; Yan et al., 2009).
In this study, we reported the cloning and characterization of the genes encoding GBSS, SSâ… , SSâ…¡, SSâ…¢ and SSâ…£ in sorghum, and the SSâ…¡b and SSâ…¢b genes in wheat. We found that the duplicator of SSâ…£ gene was lost in the sorghum, maize and wheat genomes. The expression patterns of the detected SS genes were analyzed and the evolution of the SS gene family in Gramineae was discussed.
1 Results and Discussion
1.1 Identification of the SS genes in sorghum and wheat
The previous studies reported the cloning of the genes encoding GBSSâ… , GBSSâ…¡ (Clark et al., 1991), SSâ… (Li et al., 1999b), SSâ…¡a (Li et al., 999a), SSâ…¡c (Yan et al., 2009), SSâ…¢a (Li et al., 2000), SSâ…£ (Leterrier et al., 2008) in wheat, and GBSSâ… /WX (EF089858), SSâ… (AF168786) in sorghum. In this study, we determined the complete coding domain sequences of the other six SS genes in sorghum and two in wheat. The designated gene names and GenBank accession numbers were SbGBSSâ…¡ (EF472254), SbSSâ…¡a (EU620718), SbSSâ…¡b (EU620719), SbSSâ…¡c (EU307275), SbSSâ…¢a (EU620720), SbSSâ…¢b (EU620721), TaSSâ…¡b (EU333947) and TaSSâ…¢b (EU333946). Only one SSâ…£ gene (XM_002440083) was detected in the public sorghum genomic sequences and EST database. No SSâ…£a gene was detected in wheat genomic sequences and EST database in NCBI and the raw Chinese spring genomic sequence reads using BLAST (http://www.cerealsdb.uk.net/ search_reads.htm). The domain orga- nization of SS proteins in Gramineae was listed in Table 1.
|
Table 1 Domain organization of the SS proteins in Gramineae
|
1.2 Phylogenetic relationships among the duplicated SS genes
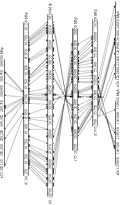
In order to characterize the identified SS genes in sorghum and wheat, a phylogenetic tree was built with 58 SS amino acid sequences from some monocots and dicots (Figure 1, a bacterium glycogen synthase, EcGS, was as out group for the phylogenetic analysis). The phylogenetic tree indicated that the SS proteins are grouped into five clades of GBSS, SSâ… , SSâ…¡, SSâ…¢, and SSâ…£. In Gramineae, GBSS proteins were further divided into two subisoforms of GBSSâ… and GBSSâ…¡, SSâ…¡ proteins into SSâ…¡a, SSâ…¡b and SSâ…¡c, SSâ…¢ proteins into SSâ…¢a and SSâ…¢b, SSâ…£ proteins into SSâ…£a and SSâ…£b, respectively (Figure 1). The present determined wheat SSâ…¡ and SSâ…¢ fall into the SSâ…¡b and SSâ…¢b subisoforms, respectively (Figure 1). The previous study found the WGD event occurred approximately 70 million years ago (Mya) prior to the divergence of the Gramineae (Paterson et al., 2004). Thus, it is likely that the duplicators of GBSS, SSâ…¡, and SSâ…¢b, but not SSâ…£, were retained in genomes of the observed Gramineae species.
|
.png) Figure 1 Phylogenetic tree derived from the full amino acid sequences of starch synthesis proteins |
1.3 Alignments of syntenic regions that contain the SSâ…£ gene among rice, maize, and sorghum genome
The two SSâ…£ genes in rice were located on chromosome 1 and chromosome 5, respectively. In order to determine whether the SSâ…£a gene is lost in sorghum and maize genomes, we compared the gene cluster on syntenic region containing the SSâ…£ genes among rice, sorghum, and maize, based on the rice-maize-sorghum synteny (http://www.gramene.org/).
The selected genomic fragment containing the SSâ…£a gene (GFa) was 544 kb on rice chromosome 1 (rC1) which contains 63 putative protein-coding genes (from Os01g0714900, a Ras-related gene, to Os01g0- 726100, an Ole e â… family gene). The genomic fragment containing the SSâ…£b gene (GFb) was 296 Kb on rice chromosome 5 (rC5) which contains 34 putative protein-coding genes (from Os05g0531200, an Ole e I family gene, to Os05g0536900, a Ras- related gene) (http://rapdb.dna.affrc.go.jp/viewer/gbr-owse/build4/) (supplement and Figure 2). There are 16 paralogous between GFa and GFb. The OsSSâ…£a gene is located between a chlorophyll a-b binding protein gene and a serine acetyltransferase gene, while the OsSSâ…£b gene located between a serine acetyltrans- ferase gene and a NADH-ubiquinone oxidoreductase 20 kD subunit gene (supplement and Figure 2).
|
Figure 2 Comparison of the starch synthesis genes on the rice/ sorghum/maize synteny physical map
|
Based on the rice-sorghum synteny (http://www.gram-ene.org/), the syntenic region of rice GFa in sorghum is 436 Kb on sorghum chromosome 3 (sC3), while the syntenic region of rice GFb in sorghum is 248 Kb on sorghum chromosome 9 (sC9). In sorghum, the GFa contains 58 putative protein-coding genes (from Sb03g032810, a Ras-related gene, to Sb03g033370, an Ole e â… family gene), while the GFb contains 36 putative protein-coding genes (from Sb09g026510, an Ole e I family gene, to Sb09g026820, a Ras-related gene) (http://rapdb.dna.affrc.go.jp/viewer/gbrowse/build4/) (supplement and Figure 2). GFa and GFb have 12 paralogous genes in sorghum. There are 44 orthologous genes in GFa, while 29 orthologous genes in GFb between in the rice genome and sorghum genome. Like the OsSSâ…£b in rice, the SbSSâ…£b gene was also located between a serine acetyltransferase 1 and a NADH-ubiquinone oxidoreductase 20 kDa subunit in sorghum. However, the SSâ…£a gene which located between the chlorophyll a-b binding protein gene and serine acetyltransferase gene in GFa in rice was lost in GFa in sorghum (supplement and Figure 2).
Based on the sorghum-maize synteny (http://www.gramene.org/), the syntenic region of sorghum GFa in maize is a 313 Kb on maize chromosome 3 (zC3), while the syntenic region of sorghum GFb in sorghum is a 64 Kb and 131 Kb on maize chromosome 6 (zC6) and chromosome 8 (zC8), respectively. In maize, the GFa contains 11 putative protein-coding genes (from GRMZM2G099166_T02, a DUF581 family protein, to GRMZM2G116282_T02, a RNA-binding protein), while the GFb on zC6 contains 6 putative protein- coding genes (from GRMZM2G033971_T01, a DUF212 family protein, to GRMZM2G043035_T02, an Ole e I family protein), and on zC8 contains 6 putative protein-coding genes (from GRMZM2G044- 866_T01, a DUF616 family protein, to GRMZM2G1- 21117_T02, a hypothetical protein) (http://rapdb.dna.affrc.go.jp/viewer/gbrowse/build4/) (supplement and Figure 2). There are 11 orthologous genes in GFa between in the rice and sorghum genome. There are 6 orthologous genes respectively in GFb on maize chromosome 6 and 8 between in the maize and sorghum genome. The ZmSSâ…£b was located between a hypothetical protein and a DUF212 family protein (supplement and Figure 2). Moreover, we can not found the SSâ…£a gene on chromosomr 3 regions in maize, and the chlorophyll a-b binding protein and the Serine acetyltransferase 3 also lost. These results strongly suggested that the SSâ…£a gene was lost in sorghum and maize genomes during their evolution.
1.4 Organ expression profile of the SS genes in sorghum and wheat
Expression divergence was often the first step in the functional divergence between duplicate genes, thus increasing the chance of retention of duplicated genes in a genome (Force et al., 1999). To define the function of the deteced SS genes, we investigated their expression in root, leaf and developing endosperm using RT-PCR in sorghum and wheat. The results indicated that the SbGBSSâ… , SbSSâ…¡a and SbSSâ…¢a genes were expressed mainly in endosperms, while the SbGBSSâ…¡, SbSSâ…¡b, and SbSSâ…¢b genes mainly in leaf in sorghum. The SbSSâ… , SbSSâ…¡c and SbSSâ…£ genes were constitutively expressed in sorghum. In addition, the SbGBSSâ…¡ and SbSSâ…¢b genes were also highly expressed in roots (Figure3A). TaSSâ…¡b and TaSSâ…¢b were mainly expressed in leaves, and moderately in roots and the early stage endosperms (Figure 3B).
|
Figure 3 The organ expression analysis of SS genes in sorghum and wheat
|
The mRNA for the TaSSâ…¡a was expressed in leaves and endosperms under the conditions used (Li et al., 1999a), but the SGP-B1 (TaSSâ…¡a) protein was not detected in leaf using monoclonal antibodies (Shimbata et al., 2005). This result is similar to the findings of OsSSâ…¡a in rice that the transcripts could be tested at a lower level in leaf, but its proteins could not be detected in leaf soluble or starch granule extracts using polyclonal antibodies (Jiang et al., 2004). The expression of TaSSâ…¡b and TaSSâ…¢b was mainly in leaves and roots, but lower in filling stage endosperms, suggests that TaSSâ…¡b and TaSSâ…¢b mainly function in leaves and roots, while TaSSâ…¡a and TaSSâ…¢a in endosperms as those in maize and rice (Jiang et al., 2004; Yan et al., 2009). These results indicated SS duplicators were also diverged in expression in wheat and sorghum as in rice and maize (Dian et al., 2003; Jiang et al., 2004; Yan et al., 2009). In addition, those SS duplication and expression divergence were similar to the starch legumes (mung bean and cowpea) genomes (Pan et al., 2009). In summary, the present study and previous reports indicate that duplicators of GBSSâ… , SSâ…¡, and SSâ…¢, but not SSâ…£, were remained in the genome and diverged in expression in all observed Gramineae species.
2 Materials and methods
2.1 Plant materials and growth conditions
The sorghum (Sorghum bicolor L.) and common wheat (Triticum aestivum L.) variety Shi4185 were planted in trail field in South China Botanical Garden, Chinese Academy of Sciences, Guangzhou, and P. R. China. The seeds were harvested four times after flowering, while the roots were got from the fifth day after germinate (DAG) seeding and full expanded leaves were taken from seedling at one day after flowering. Samples were frozen under liquid nitrogen and stored at -72℃ until use. All samples were collected in a time period between 9:00 am to 10:00 am.
2.2 cDNA cloning of GBSS, SSâ…¡, and SSâ…¢ genes in sorghum and wheat
Total RNA was isolated from sampled leaves with Plant RNAout kit (Tiandz Company, http://www.tian-dz.com). First-strand cDNA synthesis using M-MLV reverse transcriptase (Promega, http://www.promega.com) following manufacture's protocol. A specific fragment of sorghum and wheat GBSS, SSâ…¡, and SSâ…¢ genes were amplified with the primer pair (Table 2), designed based on the conserved regions of the corresponding genes from other higher plants. The purified fragments were cloned into a pMD18-T vector (TaKaRa, http://www.takara.com.cn) and confirmed by sequencing in the Invitrogen Company (http://www.invitrogen.com.cn). The 5'- and 3'-ends of sorghum GBSS, SSâ…¡, and SSâ…¢ genes were obtained with a 5' full RACE cDNA Amplification kit (Clontech) according to the user's instructions. Based on these sequences, 5’-end of the TaSSâ…¡b and TaSSâ…¢b cDNAs was determined by using the BD SMART™ RACE cDNA Amplification Kit (Invitrogen, Carlsbad, CA, USA). Putative transit peptide were identified using the ChloroP neural network analysis of the 100 amino acids at the N terminus of each sequence (http://www.cbs.dtu.dk/services/ChloroP).
|
Table 2 Primers used for sorghum in this study
|
2.3 Semi-quantitative RT-PCR analysis
Total RNA was extracted from roots, leaves and developing endosperms using the Plant RNAout kit. PCR amplifications were performed on the first cDNA strand using their specific primer sets (Table 2; Table 3). Primers (Table 2; Table 3) that amplify sorghum Actin (X79378) and wheat Actin (AY663392) were used as a control. PCR products were analyzed by 1% agarose gels, with ethidium bromide staining and take photo through Fluorescence Chemiluminescence & Visible Imaging System. Afterwards the purified fragments were cloned into the pMD18-T vector and the sequence were determined.
|
Table 3 Primers used for wheat in this study
|
2.4 Data analysis
Phylogenetic analysis was carried out using the conserved domains sequences of SS genes. Alignments of the SS sequences were aligned using the ClustalW program on EBI Web server. Phylogenetic trees were calculated using PhyML Online analysis (Guindon et al., 2005) (http://mobyle.pasteur.fr/cgi-bin/MobylePortal/portal.py?form=phyml) based on the JTT model with a constant rate of site variation (Guindon and Gascuel, 2003). Bootstrap values were calculated from 100 replicate analyses.
2.5 Gene loci comparison
To confirm their physical location, the gene loci on rice and sorghum were obtained by the alignment of the cDNA sequences from the NCBI database (http://www.ncbi.nlm.nih.gov/blast) to the corresponding chromosome-based pseudomolecules using PHYTO- ZOME blast (http://www.phytozome.net/search.php?show=blast). The location of the maize genes on the chromosome pseudomolecules was traced though the anchored corresponding BAC genome sequences or related markers (http://www.maizegdb.org). Finally, the loci of anchored rice, maize and sorghum starch synthesis genes were compared on the rice/maize/ sorghum synteny physical maps available at Gramene (http://www.gramene.org/).
Acknowledgements
This research was supported by funds received from the National Natural Science Foundation of China (No. 31070227) and the CAS/SAFEA International Partnership Program for Creative Research Teams.
References
Ball S.G., and Morel M.K., 2003, From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule, Ann. Rev. Plant Biol., 54: 207-233 doi:10.1146/annurev.arplant.54.031902.134927 PMid:14502990
Ball S.G., van de Wal M.H.B.J., and Visser R.G.F., 1998, Progress in understanding the biosynthesis of amylose, Trends Plant Sci., 3(12): 462-467 doi:10.1016/S1360-1385(98)01342-9
Brunner S., Fengler K., Morgante M., Tingey S., and Rafalsk A., 2005, Evolution of DNA sequence nonhomologies among maize inbreds, The Plant Cell, 17: 343-360 doi:10.1105/tpc.104.025627 PMid:15659640 PMCid:548811
Clark J.R., Robertson M., and Ainsworth C.C., 1991, Nucleotide sequence of a wheat (Triticum aestivum L.) cDNA clone encoding the waxy protein, Plant Mol Biol., 16(6): 1099-1101 doi:10.1007/BF00016086 PMid:1863765
Dian W.M., Jiang H.W., Chen Q.S., Liu F.Y., and Wu P., 2003, Cloning and characteriza- tion of the granule-bound starch synthase II gene in rice: Gene expression is regulated by the nitrogen level, sugar and circadian rhythm, Planta, 218(2): 261-268 doi:10.1007/s00425-003-1101-9 PMid:12955512
Dian W.M., Jiang H.W., and Wu P., 2005, Evolution and expression analysis of starch synthase III and IV in rice, J. Exp. Bot., 56(412): 623-632 doi:10.1093/jxb/eri065 PMid:15642712
Force A., Lynch M., Pickett F.B., Amores A., Yan Y.L., and Postlethwait J., 1999, Preservation of duplicate genes by complementary, degenerative mutations, Genetics, 151: 1531-1545 PMid:10101175 PMCid:1460548
Gaut B.S., and Doebley J.F., 1997, DNA sequence evidence for the segmental allotetraploid origin of maize, Proc. Natl. Acad. Sci., USA, 94(13): 6809-6814 doi:10.1073/pnas.94.13.6809
Guindon S., and Gascuel O., 2003, A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood, Syst Biol., 52(5): 696-704 doi:10.1080/10635150390235520 PMid:14530136
Guindon S., Lethiec F., Duroux P., and Gascuel O., 2005, PHYML Online a web server for fast maximum likelihood-based phylogenetic inference, Nucleic Acids Res., 33: 557-559 doi:10.1093/nar/gki352 PMid:15980534 PMCid:1160113
Harn C., Knight M., Ramakrishnan A., Guan H., Keeling P.L., and Wasserman B.P., 1998, Isolation and characterization of the zSSIIa and zSSIIb starch synthase cDNA clones from maize endosperm, Plant Mol. Biol., 37(4): 639-649 doi:10.1023/A:1006079009072 PMid:9687068
Hirose T., and Terao T., 2004, A comprehensive expression analysis of the starch synthase gene family in rice (Oryza sativa L.), Planta, 220(1): 9-16 doi:10.1007/s00425-004-1314-6 PMid:15232694
Jiang H.W., Dian W.M., Liu F.V., and Wu P., 2004, Molecular cloning and expression analysis of three genes encoding starch synthase II in rice, Planta, 218(6): 1062-1070 doi:10.1007/s00425-003-1189-y PMid:14740212
Leterrier M., Holappa L.D., Broglie K.E., and Beckles D.M., 2008, Cloning, characterisa- tion and comparative analysis of a starch synthase IV gene in wheat: functional and evolutionary implications, BMC Plant Biol., 8: 98
doi:10.1186/1471-2229-8-98 PMid:18826586 PMCid:2576272
Li Z.Y., Chu X.S., Mouille G., Yan L.L., Kosar-Hashemi B., Hey S., Napier J., Shewry P., Clarke B., Appels R., Morell M.K., and Rahman S., 1999a, The localization and expression of the class II starch synthases of wheat, Plant Physiol., 120: 1147-1156 doi:10.1104/pp.120.4.1147 PMid:10444098 PMCid:59348
Li Z.Y., Mouille G., Kosar-Hashemi B., Rahman S., Clarke B., Gale K.R., Appels R., and Morell M.K., 2000, The structure and expression of the wheat starch synthase III gene: motifs in the expressed gene define the lineage of the starch synthase III gene family, Plant Physiol., 123(2): 613-624 doi:10.1104/pp.123.2.613 PMid:10859191 PMCid:59029
Li Z.Y., Rahman S., Kosar-Hashemi B., Mouille G., Appels R., and Morell M.K., 1999b, Cloning and characterization of a gene encoding wheat starch synthase I, Theor Appl Genet., 98(8): 1208-1216 doi:10.1007/s001220051186
Pan X.X., Tang Y.Y., Li M.R., Wu G.J., and Jiang H.W., 2009, Isoforms of GBSSI and SSII in four legumes and their phylogenetic relationship to their orthologs from other angiosperms, J Mol Evol., 69(6): 625-634
doi:10.1007/s00239-009-9300-z PMid:19888543
Paterson A.H., Bowers J.E., and Chapman B.A., 2004, Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics, Proc Natl Acad Sci USA, 101(26): 9903-9908
doi:10.1073/pnas.0307901101 PMid:15161969 PMCid:470771
Roldán I., Wattebled F., Mercedes L.M., Delvallé D., Planchot V., Jiménez S., Pérez R., Ball S., D'Hulst C., and Mérida A., 2007, The phenotype of soluble starch synthase IV defective mutants of Arabidopsis thaliana suggests a novel function of elongation enzymes in the control of starch granule formation, Plant J., 49(3): 492-504 doi:10.1111/j.1365-313X.2006.02968.x PMid:17217470
Shimbata T., Nakamura T., Vrinten P., Saito M., Yonemaru J., Seto Y., and Yasuda H., 2005, Mutations in wheat starch synthase II genes and PCR-based selection of a SGP-1 null line, Theor. Appl. Genet., 111(6): 1072-1079
doi:10.1007/s00122-005-0032-1 PMid:16172895
Smith A.M., Denyer K., and Martin C., 1997, The synthesis of the starch granule, Annu Rev Plant Physiol Plant Mol. Biol., 48: 67-87 doi:10.1146/annurev.arplant.48.1.67 PMid:15012257
Song R., and Messing J., 2003, Gene expression of a gene family in maize based on noncollinear haplotypes, Proc. Natl. Acad. Sci., USA, 100(15): 9055-9060 doi:10.1073/pnas.1032999100 PMid:12853580 PMCid:166437
Vrinten P.L., and Nakamura T., 2000, Wheat granule-bound starch synthase I and II are encoded by separate genes that are expressed in different tissues, Plant Physiol., 122(1): 255-264 doi:10.1104/pp.122.1.255 PMid:10631269 PMCid:58864
Wendel J.F., 2000, Genome evolution in polyploids, Plant Mol. Biol., 42(1): 225-249 doi:10.1023/A:1006392424384 PMid:10688139
Yan H.B., Jiang H.W., Pan X.X., Li M.R., Chen Y.P., and Wu G.J., 2009, The gene encoding starch synthase IIc exists in maize and wheat, Plant Science, 176(1): 51-57 doi:10.1016/j.plantsci.2008.09.003
Yan H.B., Pan X.X., Jiang H.W., and Wu G.J., 2009, Comparison of the starch synthesis genes between maize and rice: Copies, chromosome location and expression divergence, Theor. Appl. Genet., 119(5): 815-825
doi:10.1007/s00122-009-1091-5 PMid:19593540
. PDF(440KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Xiaoxue Pan
. Hongbo Yan
. Meiru Li
. Guojiang Wu
. Huawu Jiang
Related articles
. Starch synthesis
. Gene duplication
. Gene divergence
. Sorghum ( Sorghum bicolor L.)
. Common wheat ( Triticum aestivum L. )
Tools
. Email to a friend
. Post a comment
.png)

.png)
.png)


