Integration and Expression Stability of Transgenes in Hybriding Transmission of Transgenic Rice Plants Produced by Particle Bombardment 
2 State Key Laboratory of Rice Biology, China National Rice Research Institute, Hangzhou, 310006, China;
3 College of Life and Environmental Science, Hangzhou Normal University, Hangzhou, 310018, China
 Author
Author  Correspondence author
Correspondence author
Molecular Plant Breeding, 2011, Vol. 2, No. 8 doi: 10.5376/mpb.2011.02.0008
Received: 26 Apr., 2011 Accepted: 03 May, 2011 Published: 20 Jun., 2011
Zhao et al., 2011, Integration and Expression Stability of Transgenes in Hybriding Transmission of Transgenic Rice Plants Produced by Particle Bombardment Vol.2 No.12 (doi: 10.5376/mpb.2011.02.0008)
Four transgenic rice lines TR 5, TR 6, Ming B and Jingyin 119 obtained via particle bombardment were used as transgene donors to create hybrids. The integration and expression stability of exotic bar and cecropin B gene in conventional hybriding transmission were investigated by Southern and Northern blotting analyses. The selection marker bar gene was transferred to all hybrids under selection of Basta herbicide. Loss or gain of small hybridization bands (no more than 2.0 kb) of bar gene occurred in some hybrids, but the difference in integration sites of bar gene copies did not influence their stable expression. The non-selection gene cecropin B was stably transferred from the four transgene donors to their resulting hybrids, but expression level was very different. Silencing of cecropin B gene occurred in some hybrids from TR 5, TR 6 and Ming B. In the transgene donor Jingyin 119 and all its resulting hybrids, cecropin B and bar gene were stably expressed. We concluded that the stability of transgene during crossbreeding transmission is mainly determined by the primary transgenic donors and may be affected by recombination.
Rice is the world’s most important food crop and a primary source of food for more than half the world’s population (Khush, 2005). Improvements in crop yield have been achieved through conventional hybridization and selection procedures (Peng et al., 2007). Currently, rice hybrids are cultivated in about 55% of the rice-growing areas in China and contribute to 66% of the total rice production of the whole country (Wu and Luo, 2007). The breakthrough in plant biotechnology has provided tools to develop more elite transgenic donors for crop crossbreeding. In the transformation process, ideally, a single gene with its appropriate controlling elements is added to the genome instead of moving a whole chromosome or chromosome segment with many known and unknown genes as done with the traditional hybridizations (Horvath et al., 2001). Since protocols for rice transformation have been well established, genetic transformation is currently complementing conventional breeding programs in the development of advanced germplasm (Popelka and Altpeter, 2003). In 1996, the herbicide-resistant bar gene was transformed into Japonica rice Jingyin 119 via particle bombardment (Huang et al.,1996), and then the transgenic plants were successfully crossed to the male-sterile line Pei-ai 64S of a two-line crossbreeding system for creating transgenic rice hybrids. Based on the herbicide resistance of the marker gene bar, a new method was created to examine and improve the purity of hybrid rice (Huang et al., 1998). More recently, Chen et al. introduced the synthetic cryl2A* and crylC* gene into an elite Indica cytoplasm male-sterile restorer line Minghui 63 and crossed the transgenic lines to Zhenshan 97A to produce insect-resistant hybrid rice (Chen et al., 2005; Tang and Lin, 2007). Transgenic technology is providing new tools for the conventional crossbreeding of rice.
The main methods used for rice transformation are particle bombardment and Agrobacterium-mediated transformation. Although the generation of transgenic plants is relatively easy for many rice varieties, the transformation frequency is usually low and rather genotype-dependent. Also, these gene delivery techniques need undergo the obligatory processes of tissue culture, which often results in phenotypic abnormalities and reduced fertility of the transgenic plants obtained (Zhang et al., 2005). Like mutants providing useful traits, transgenic plants often have to be used for relocating the gene in more suitable genotypes (Horvath et al., 2001). Thus, the success of plant genetic manipulation not only requires the stable inheritance and expression of transgenes in the transgenic plants across generations, but also depends on whether the transgenic plant can be used as a transgene donor in recombination crossbreeding.
Many studies have analysesed the progenies of the primary rice transformants, revealing that transgene stability was significantly related to differences in transgene structure and expression levels between transgenic lines, particularly in transgenic plants derived from direct DNA transfer such as particle bombardment (Vain et al., 2002; Altpeter et al., 2005). In transgenic cereals, more than 50% of transgenes can be inactivated over successive generations (Iyer et al., 2000). These problems make molecular genetic studies difficult, and frustrate attempts at crop improvement through genetic engineering. Additionally, they create difficulties in predicting transgene behavior when transgene needs to be transferred by conventional crossing (Vain et al., 2002). Altpeter et al. (2005) speculated that particle bombardment might be advantageous over Agrobacterium-mediated transformation in respect of transferring the transgenes into a new genetic background via traditional breeding, because by particle bombardment multiple transgenes are tend to be integrated into the same locus . But there are few direct evidences for this question up to date.
We introduced the plasmid pCB1 carrying the selected herbicide-resistant bar gene and the non-selected cecropin B gene into four Japonica rice varieties via particle bombardment between 1996 and 1998. Bar gene was introduced into rice plant for resistance to phosphinothricin (the active component of the herbicide Basta) and the cecropin B gene was used to resist a range of plant pathogenic bacteria including Xanthmomonas compestris pv oryzae, which leads to rice leaf bacterial blight disease. With obvious phenotype and convenient detection, bar gene has been proved to be a very useful marker to screen transgenic hybrids. In the past ten years, the elite transgenic rice plants harboring bar and cecropin B gene were selected as transgene donors to cross to different rice varieties. We constructed a population of rice hybrids derived from multiple conventional crosses. Here we report the inheritance and expression behaviours of the foreign bar and cecropin B genes during rice crossbreeding transfer.
1 Results and Analysis
1.1 Stability of transgene integration patterns in mono-cross transmission
The stability of integration patterns for the selected bar gene and non-selected cecropin B gene in the transmission from transgenic donors to hybrid rice plants was investigated using genomic DNA Southern blotting analysis. Three transgene donors including TR 5, TR 6, Ming B were used to produce hybrids, in which several bar gene copies were inherited as a single transgenic locus when tested by Basta resistance (Hua et al., 2003).The transgene integration patterns of transgene donor TR 5, TR 6, Ming B and their corresponding hybrids were analysed and shown in Figure 1A and 1B. Southern blotting results revealed as follows: (i) The tightly linked bar and cecropin B gene in the original plasmid exhibited different integration patterns in the three transgene donors, when hybridized with bar and cecropin B probe respectively, after genomic DNA was digested with Hind â…¢, which cut once in plasmid pCB1. There were two hybridization bands of bar gene and three bands of cecropin B gene in transgene donor TR 5. Transgene donor Ming B had four bar gene hybridization bands and three cecropin B gene hybridization bands. TR 6 donor plant possessed six hybridization bands of bar gene and five bands of cecropin B gene. These indicated that bar and cecropin B might have different copies integrated in the receptor genomes. (ii) In the self-pollinated progenies of transgenic rice hybrids, the integration patterns of non-selected cecropin B gene remained the same as that of their corresponding transgene donors (Figure 1B). However, the integration patterns of bar gene were changed in some hybrids (Figure 1A). For example, two bands of 1.5 kb and 2.0 kb in length were lost from the progeny plants of cross lines TR 6/CJN 2 and TR 6/Bing 95-13, comparing with their transgene donor TR 6. Two new hybridization bands of bar gene, 1.5 kb and 2.0 kb in length, emerged in the progeny plant of cross line TR5/CJN 3, comparing with its transgene donor TR5. Although the TR5 lane was loaded with less DNA, giving possibility of the 1.5 kb and 2.0 kb bands of bar gene coming from its parent TR5, these two new hybridiztion bands did not appear in the remaining other three hybrids from TR5, whoes lanes were loaded with more DNA (Figure 1A). These confirmed the conclusion that the 1.5 kb and 2.0 kb hybridization bands of bar gene were created in cross line TR5/CJN 3.
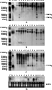 Figure 1 Integration and expression analyses of selected bar and non-selected cecropin B gene in rice in mono crossing transmission |
1.2 Stability of transgene integration patterns in multiple crosses transmission
The inheritance of transgene bar and cecropin B in the course of multiple crosses was revealed by DNA Southern blotting analysis, using Jingyin 119 line as transgene donor. This Jingyin 119 transgenic line had four bar gene loci and two cecropin B gene loci when hybridized with their probe respectively, after genomic DNA was digested with Hind â…¢, which cut once in the plasmid pCB1 (Figure 2A and 2B). Southern blotting results demonstrated that the integration pattern of non-selected cecropin B gene was very stable in mono- and multiple crossbreeding transmission (Figure 2B). All the progenies of hybrids showed two hybridization bands of cecropin B gene, exactly the same as that of Jingyin 119 donor. But the integration pattern of selected bar gene did not always remain stable during crossbreeding transmission. Among the seventeen rice crosses, the integration pattern of bar gene remained stable in eight hybrids and changed in the other nine lines, which lost two smaller hybridization bands of bar gene, 1.6 kb and 1.0 kb in length respectively (Figure 2A). Our former research revealed that the transgenic integration patterns of Jingyin 119 transgene donor kept stable in self-pollination across generations, but bar and cecropin B gene showed different integration patterns when Southern-blotting analyses were conducted after genomic DNA digested with different enzymes (Hua et al., 2003). During crossing transmission the two smaller hybridization bands of bar gene were lost in some hybrids while the other two bigger bands were transmitted stably (Figure 2A and 2B).
We also noticed that disappearance of the two smaller fragments (1.6 kb and 1.0 kb) of bar gene did not depend on the cross turns (Figure 2A). For example, the cross hybrid of 119/Bing 94-02 kept the same integration pattern of bar gene as that of its transgene donor Jingyin 119, but in its related re-cross hybrid Jingyin 119/Bing 94-02//T951, the 1.6 kb and 1.0 kb hybridization bands of bar gene were lost. Another hybrid rice line of cross Jingyin 119/59 lost the two smaller fragments of bar gene, but in progenies of its four related multiple crossing hybrids, three crossing combinations including Jingyin 119/59//L97-55, Jingyin 119/59//DS4 and Jingyin 119/59//DS4///Jingyin 119/31//9522 lost the two smaller fragments of bar gene, the remaining one cross Jingyin 119/59//T951 exhibited the same bar gene integration pattern as that of the original Jingyin 119 donor. There was also one hybrid line Jingyin 119/57//DS4///L97-55 that carrying all the original four bar gene loci of its transgene donor after three crossing turns. These suggested that loss of bar gene fragments in transmission of multiple crosses was not related to crossing turns, but mainly depended on the primary hybrid plants that were randomly selected as transgene donors for the next crosses.
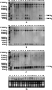 Figure 2 Integration and expression analyses of selected bar and non-selected cecropin B gene in rice in multiple crossing transmission |
Moreover, disappearance of the bar gene smaller fragments occurred in both hybrids when transgene donor Jingyin 119 was male parent (C20/Jingyin 119) and female parent (e.g. Jingyin 119/59). This demonstrated the bar gene smaller fragments harbouring in donor plant had equal chance to be lost through pollen and egg. Therefore, we speculated that the two smaller hybridization bands of bar gene in Jingyin 119 transgene donor might integrate in one separate genomic locus from the other bigger ones and more possibly, possess no expression activity.
1.3 Stability of transgene expression in crossing transmission
During the course of crossbreeding for generating different transgenic hybrid rice lines, all the hybrids were subject to Basta-resistance assay and only the resistant plants were selected. These ensured the existence and expression of selected bar gene in all hybrids. What about the fate of the non-selected cecropin B gene? The completeness of cecropin B gene expression cassette in hybrid rice lines was examined by Southern blotting analyses and the expression status of cecropin B in crossing transmission was revealed by Northern blotting analyses.
To detect the completeness of cecropin B gene expression cassette in rice genome, Southern blot was conducted after genomic DNA was digested with Pstâ… and Hindâ…¢, which released a 1.12 kb fragment comprising of the coding region of cecropin B gene and its pin terminator (Figure 3). Results showed that presence of the predicted 1.12 kb fragment in hybrid lines mainly depended on their original transgenic donors (Figure 1C and 2C). Transgene donor TR 5, TR 6 and their derived hybrids did not exhibit the expected 1.12 kb fragment, illustrating there was no intact cecropin B copies, or more probably, the cut sites of restriction enzymes (Pst â… and Hind â…¢) in cecropin B gene expression cassette were modified. Transgene donor Ming B, Jingyin 119 and their hybrids generated the 1.12 kb fragment as expected, revealing that there was at least one intact copy of cecropin B gene in these transgenic lines.
| Figure 3 The probe structure and site on pCB1 |
Northern blot results showed the expression behaviour of non-selected cecropin B gene varied significantly among transgenic donors and their hybrid lines (Figure 1D and Figure 2D). The expression of cecropin B gene in the primary transformants (T0 generation) of all the four transgene donors including TR 5, Ming B, TR 6 and Jingyin 119 was proved by Northern blot analysis (data not shown). However, in their self-pollinated offspring, gene silence of cecrropin B occurred in TR 5 and Ming B donor (Figure 1D), both harbouring 3 hybridization bands of cecropin B (Figure 1B). The TR 6 and Jingyin 119 donors, with 5 and 2 hybridization bands of cecropin B gene respectively (Figure 1B and 2B), expressed cecropin B gene stably arcoss 6 and 12 generations, respectively. This indicated that silencing of non-selected cecropin B over generations was relevant to different transformation events rather than the number of integrated hybridization bands. In view of the Southern blot results, we also concluded that there was no certain relationship between the cecropin B gene expression status and the emergence of the expected 1.12 kb fragment, which was used to predict the completeness of cecropin B gene copy.
The expression behaviour of cecropin B gene in cross transmission was revealed by comparing the Northern blot results of transgenic hybrids with that of their corresponding donors. Gene silence of cecropin B occurred in both self-pollinated progeny of TR 5 donor (T5) and all its derived hybrid lines (F3) (Figure 1D). In TR 6 transgene donor, cecropin B gene was expressed at mRNA level over 6 generations, but silenced in its two hybrid lines (F3 generation). Interestingly, cecropin B gene did not express in the selfed progenies (T6) of transgene donor Ming B, but expressed in its two out of three cross lines Ming B/Jia 59 and Ming B/Xuzao, till F3 generation (Figure 1D). The stable expression of cecropin B gene was observed in Jinyin 119 donor and all its hybrids, ignoring the complex cross combinations and multiple cross turns. We analysed the cecropin B expression in the progeny plants of 14 out of 17 hybrids from Jingyin 119 donor (Figure 2D). Whether transgnene donor Jinyin 119 was female parent or male parent (C20 / Jingyin 119), whether transgenes in Jingyin 119 were transferred through one cross turn (Jingyin 119/57, Jingyin 119/Bing 94-02, Jingyin 119/59), two cross turns (Jingyin 119/59//L97-55, Jingyin 119/57//9522, Jingyin 119/390//S1, Jingyin 119/63//T951, Jingyin 119/Bing 94-02//T951, Jingyin 119/02//T951, Jingyin 119/63//390, Jingyin 119/59//DS4 and Jingyin 119/503//T951) or three cross turns (Jingyin 119/ 57 // DS4 /// L97-55), or stacked by crossing between transgenic hybrids (Jingyin 119 / 59 // DS4 /// Jingyin 119 / 31 // 9522), the non-selected cecropin B gene expressed stably in all hybrids over 6 to 8 generations. In general,the expression behaviour of non-selected cecropin B gene was complex in crossbreeding transmission,depending on its integration structures as well as the genetic background of host plants.
1.4 Heredity and expression behavior of transgenes in F1 hybrid of transgene donors
The heredity and expression behaviour of the exotic bar and cecropin B gene in F1 hybrid of transgene donor TR 5 (T5 generation) and TR 6 (T6 generation)was investigated. Southern-blot results showed that in F1 hybrid of TR 5/TR 6, bar and cecropin B gene exhibited the complementary integration patterns to that of their parents (Figure 1A and 1B), suggesting transgenic integration locations in TR 5 and TR 6 were non-allelic. But a 2.0 kb hybridization band of bar gene from TR 6 parent was lost, further confirming the instability transmission of selected bar gene. As to the expression of transgenes, the F1 hybrid rice plant exhibited Basta-resistance, implying the normal expression of selectable bar gene. However, the non-selected cecropin B gene was only expressed in TR 6 parent plant and silenced both in TR 5 parent and the F1 hybrid (Figure 1D). It seemed that the silencing status of cecropin B in TR 5 could exert negative effect on cecropin B gene expression in F1 hybrid.
2 Discussion
2.1 Stability of transgene integration patterns in crossing transmission
Whether transgenes could be transferred from transgenic donor plants into other varieties through conventional crossbreeding is crucial for the successful application of transgenic technique in plant breeding. Present results described the stability of integration patterns and expression activities of transgenes in rice during the course of crossbreeding transmission.
We found in process of crossing transfer, the integration patterns of non-selected cecropin B gene were very stable. All the investigated hybrids kept the same cecropin B integration patterns as that of their corresponding donor plants. However, the integration patterns of selected bar gene showed instability to certain degree. One obvious phenomenon is gene losing. The lost hybridization bands of bar gene had relatively smaller molecular weight (no more than 2.0 kb), usually companying with the stable transfer of two or more larger Southern hybridization bands. These indicated that the stably transferred larger bands of bar gene were expressed and mainly provided the Basta-resistance for selection of transgenic hybrids. The smaller bands of bar gene that tended to be lost in crossing transmission might integrate in different chromosomal location from the larger ones. In the course of crosses, these smaller bands were transferred as unlinked (as in TR 6) or partly-linked (as in Jingyin 119) unit with the larger bands of bar gene, and most probably possessed no expression activity, as there was no hybrids harbouring the smaller hybridization bands of bar gene only. We speculated that when these smaller bands of bar gene happened to be caught by hybrid plants independently, the hybrid plants could not survive because every hybrid in our experiments was subjected to Basta-resistance screening.
Also, there is the possibility that the loss of bar gene fragments was aroused from DNA recombination events involved in transgenic loci. Gene loss from one generation to the next was previously reported in maize (Register et al.,1994) and rice (Vain et al., 2002). Some transgenic loci generated by particle bombardment are likely to be altered from one generation to the next through recombination or deletion (Vain et al., 2002). Molecular studies have shown that direct DNA transfer often leads to integration at one locus of multiple fragmented and rearranged transgene copies as well as plasmid backbone sequences (Kohli et al., 2003). In the four transgene donor plants, differences between the integration patterns of bar and cecropin B gene were investigated, although they were tightly linked in the plasmid construction. These illustrated that rearrangement or recombination might occur between bar and cecropin B gene cassettes during their integration. The 19 bp palindromic sequence within TATA-box of the CaMV35S promoter is proved to be a recombination hotspot that can confirm the predominance of microhomology-mediated recombination of plasmid rearrangement in transgenic integration (Kohli et al., 1999). Bar gene in present research is driven by CaMV35S promoter. Thus the recombination triggering sequences in CaMV35S promoter not only promoted the plasmid rearrangements during integration, but might exert effects on the DNA recombination events of bar gene loci in the subsequent crossing transmission. In deed, the presence of new transgenic fragments of bar gene in hybrid TR 5/CJN 3 further confirmed the rearrangement of bar gene integration loci. More importantly, we found that the variance existing in integration patterns of bar gene did not interfere with its stable expression and the successful selection of the corresponding hybrids. This indicated that transgenes introduced by particle bombardment could be successfully transferred in conventional crossbreeding.
2.2 Stability of transgene expression in crossing transmission
Different integration sites, copy numbers and transgenic locus configurations, as well as epigenetic silencing mechanisms are revealed to be the main factors influencing transgene expression by previous researchers (Iyer et al., 2000; Meyer, 1995; Matzke and Matzke, 1998). However, documents on the relationship between transgene expression and crossing transmission are very limited up to date. Several reports described the expression behaviour of transgenes in rice crossbreeding in recent years, which giving the opinion that at least the same transgene expression level in hybrids, if no more than that of the transgenic parents, could be expected (Chen et al.,2005; Tang et al., 2006; 2007; Wang and Lin, 2007). But the above results were established on the base of transgenic donors containing single-copy exotic gene and transformed by Agrobacterium tumefaciens method. Our results revealed the different fate of transgenes in crossbreeding transmission that was introduced by DNA direct delivery system.
The selected bar gene is expressed in all hybrids and their corresponding transgenic parents across generations in spite of its variability in integration patterns. However, the co-expression behaviour of non-selected cecropin B gene was very complex. Both inactivation (e.g. in hybrids of TR 6) and maintaining of successive expression activity (e.g. in hybrids of Ming B) of cecropin B gene were observed through crossing transmission. The stable expression of cecropin B gene were also observed after kinds of cross combinations across several generations (e.g. in hybrids of Jingyin 119 donor). We confirmed that expression status of selected bar and non-selected cecropin B was independent within the same genomic locations. This means the inactivation of cecropin B does not spread over to the adjacent bar gene, which is consistent with the conclusion of some previous reports (Vain et al., 2002; Kohli et al., 2003) but argues against others (Lindsay et al., 1996).
As expression status of non-selected cecropin B gene was very different among the four transgenic donors in crossing transfer, we concluded that the structure of transgenic loci and the chromosomal locations where transgenes integrated are main factors influencing gene expression in sexual reproduction as well as in conventional crossbreeding. TR 5 is a typical transgenic line that non-selected cecropin B gene expressed in primary transformant but silenced in progenies of both self-pollination and crosses. This indicated the integration structure and/or transgenic loci of cecropin B in TR 5 are prone to trigger gene silence. The instability of selected bar gene integration pattern in TR 5 hybrids illustrated that transgenic loci of TR 5 line were recombination triggering. Silencing of transgene is often aroused by homologous sequences between multiple transgene copies or between exotic DNA and endogenous DNA sequences of the host plant, either the homologous sequences at allelic or nonallelic chromosomal locations(Kumpatla et al.,1997). It is probable that homologous sequences of different copy or fragment of cecropin B gene caused its own inactivation. The possibility of interactions between actin promoter and endogenous homologous sequences in rice genomic DNA could not be ruled out, for the coding region of cecropin B gene is driven by actin promoter from rice. Homology-mediated gene silencing is based on DNA-DNA pairing, which might be involved in the case where longer time and more generations are needed. It could be strongly triggered when transgenes are arranged in palindromic manner or in inverted repeat (IR) (Fojtová et al., 2006). Whether IR exists in transgenic loci of TR 5 needs to be revealed by extensive research on the sequence of transgenic loci in this line.
Northern blotting analysis showed the inactivation of cecropin B gene occurred at transcriptional level. Transcriptional gene silencing in plant is often associated with DNA methylation in the promoter region and the 5’ un-translated region. DNA methylatin-correlated gene inactivation can be released after treatment with methylation inhibitors such as 5-azacytidine (Kohli et al., 1999; Kumpatla et al., 1997). Further experiments need to be conducted for investigating whether DNA methylation is involved in the cecropin B gene silence.
An interesting result of the present study is that we found Jingyin 119 is a special transgenic line for transgenes to be expressed stably. Both cecropin B and bar gene were stably expressed in Jingyin 119 donor and all its resulting hybrids, ignoring the complex crossbreeding transmission and across generations. Previously, the stable inheritance and expression of exotic bar and cecropin B gene in this Jingyin 119 transgenic line was confirmed over six successive generations (Kumpatla et al., 2001). Here we presented the foreign bar and cecropin B gene in this transgenic line kept expressing across long-term generations of self-pollination (T12) and after multiple crossing transmission. It is interesting to inquire into the following questions: is the integration structure of exotic bar and cecropin B in Jingyin 119 donor very suitable for their expression? Are there safe chromosomal locations in rice genome for transgenes to reside and escape from the rice genome supervision and modification system, just as the locations in Jingyin 119 genome where exotic bar and cecropin B integrated? Meyer (1995) proposed that endogenous, transcriptional active sequences contain cis-acting flanking regions are necessary for adequate functioning of genetic machinery, for these sequences can facilitate DNA bending or elicit different trans-acting factors. Such ‘isochore’ sequences are special sequences. Any deviation from isochore structure is recognized as a foreign element, which may lead to elimination or silencing of transgene. Thus, how and why foreign bar and cecropin B in Jingyin 119 could escape from the recognition and attack of the rice genome defending system? Information regarding the chromosomal locations favouring transgene expression is very limited so far. The excellent performance of transgenes in Jingyin 119 transgenic line provides a promising hint to seek for these suitable chromosomal locations for transgene integration. Further experiments are necessary to discover the transgene structure and integration positions on rice chromosome of bar and cecropin B gene in Jingyin 119 transgenic.
Another phenomenon we found in present study is that conventional crossing transmission can affect the expression status of non-selected cecropin B gene. The inactivation of cecropin B gene in crosses of TR 6 and the maintaining successive expression activity of cecropin B gene in some crosses of Ming B implied that effect from host genotypic constitution on transgene expression can not be ignored. Under these conditions, the effects from integration position, copy number and rearrangement of transgenic loci on cecropin B expression could be ruled out, because the integration patterns of cecropin B gene in hybrids are exactly the same to their corresponding transgenic donors. The mechanism to explain how host genotypic constitution influenced transgene expression is possible at epigenetic level, such as change of methylation status of cecropin B gene expression cassette. The alteration in genotype backgrounds by conventional crosses might prevent, or at least delay, the epigenetic modification process of transgenes in some circumstances, which were illustrated by the expression activity of cecropin B in some hybrids of Ming B.
On the other hand, we used the progenies of transgenic donors and hybrids as materials to investigate the transgene expression behaviour in crossing transmission. It remains unclear whether transgene inactivation in hybrids occurred immediately after cross (in F1 generation) or through generations. The results of F1 hybrid of TR 5/TR 6 may throw some light on this question. Cecropin B gene silenced in TR 5 donor but expressed normally in TR 6 donor. The subsequent F1 hybrid of TR 5/TR 6 lost its express activity of cecropin B (Figure 1D). This implied the inactivation of cecropin B gene occurred immediately after cross. Fojtová et al (2006) revealed in tobacco that meiosis could not alter the expression and methylation patterns established in the hybrid plants. Thus we speculated the inactivation of cecropin B gene occurred in F1 hybrids and the silencing status was passed on through sexual reproduction.
In conclusion, transgenes in rice genome introduced by particle bombardment could be easily transferred by conventional crossbreeding. The stability of transgene integration patterns and expression status in crossbreeding transmission was mainly determined by the primary transgenic donor plant. In the course of cross transmission, the variance in integration patterns of selected marker bar gene did not influence its stable expression under selection, whereas the expression status of non-selected cecropin B gene is affected by cross recombination. And more importantly, transgenic plant can be produced by particle bombardment in which transgenes sustains inheritance and expression activity over long-term generations in both sexual reproduction and conventional crossbreeding.
3 Materials and methods
3.1 Generation of transgenic plants
Four transgenic rice lines TR 5, TR 6, Ming B and Jingyin 119 transformed via particle bombardment were used as transgene donors to create transgenic hybrids. All the above transgenic donors were generated as described in previous reports, harboring plasmid pCB1, which contains the selected marker-gene bar and non-selected cecropin B gene. The bar gene is controlled by cauliflower mosaic virus 35S (CaMV 35S) promoter, cecropin B gene is controlled by the rice actin-1 promoter (Huang et al., 1996).
3.2 Crosses
The transgenic plants of each rice variety were identified by both Basta-resistant phenotype analyses and Southern-blotting hybridization. Bagged seeds of transgenic T0 plants of Jingyin 119, TR 5, TR 6 and Ming B variety were planted in the experimental farm of China National Rice Research Institute between 1996 and 1998. Seeds of each plant were harvested individually and the elite homozygous transgenic lines were selected as transgene donors for cross experiments.
All cross experiments were conducted in the field in the rice growing seasons between 1996 and 2009 both in Zhejiang Province and Hainan Province. A cross between transgene donor TR 5 (T5) and TR 6 (T6) was made to investigate the relationship between integration and expression of transgenes in different transgene donors. The homozygous transgenic line Jingyin 119 (T3 generation) was used as transgene donor to cross with different Japonica rice varieties. Some of the resulting F1 hybrids were used as new transgene donors for next crosses to produce hybrids of multiple crosses. All the selected transgene donors and hybrids were subjected to Basta resistant assay in every generation. We sprayed 0.32% Basta to whole plant at their seedling stage. After one week, the susceptible plants were dead. The selected transgenic plants were grown in the field for seeding or cross experiment. The transgenic hybrids were self-crossed to produce homozygotes for the bar gene, and progenies were selected based on Basta resistance and Southern hybridization. All homozygous transgenic hybrids and their transgenic donors were planted in the experimental farm of China National Rice Research Institute. The information of rice materials for experiments was detailed in Table 1.
.png) Table 1 Pedigree of transgenic rice cross lines used for transgene integration and expression analyses |
3.3 Southern blot hybridization
Genomic DNA was isolated from rice leaves using the SDS DNA extraction method as described by Lu and Zheng (1992) Aliquots (5 μg) were digested overnight with appropriate restriction enzymes, fractionated by 0.8% agarose gel electrophoresis and blotted onto Amersham N+ Hybond membranes according to the Southern blotting method (Sambrook et al., 1989). The linear fragment (0.9 kb) comprising the open reading frame of bar gene and nos terminator from pCB1 digested with EcoRV was used as probe for bar gene. The linear fragment (1.12 kb) from pCB1 after digestion with Hindâ…¢ and Pstâ… , which comprises cecropin B coding region and Pin terminator was used as probe for cecropin B gene. The probe structure and sites on pCB1 are illustrated in Figure 3.
The probe labeled by random priming was conducted using DIG DNA Labeling Kit (Roche Company) according to the manufacture’s instructions. Southern blot hybridization and detection was carried out using DIG Luminescent Detection Kit (Roche Company) according to the manufacture’s instructions.
3.4 Northern blot hybridization
Leaf tissue (0.5-1g) was ground in liquid nitrogen. The dispersed tissue was used for total RNA extraction using TRIzol Kit (Invitrogen Company) according to the manufacture’s instructions. Extracted RNA was initially checked for quality and quantity on normal 1% agarose gel. After electrophoresis on 1.2% formaldehyde-agrose gel, the gel was washed for 10 min in sterile water to remove the formaldehyde. The RNA was denatured in 0.05 mol L-1 NaOH and blotted onto Amersham N+ Hybond membranes in 20× SSC according to the Southern blotting method(Meyer et al., 1995). The DNA probe of cecropin B gene described as above was used for Northern hybridization.
Northern blot hybridization and detection was carried out using DIG Luminescent Detection Kit (Roche Company) according to the manufacture’s instructions.
Authors’ contributions
Yan Zhao conducted the major part of this study including experimental design, Southern and Northern hybridization experiment, and manuscript preparation. Longbiao Guo and Huizhong Wang conducted the rice material hybrid design and the crossing experiment. Danian Huang participated in experimental design.
Acknowlegedments
This work was supported by grants from the National Nature Science Foundation of China (No: 30871511&30771317), the key program from the Ministry of Agriculture of China for Creation of New Transgenic Organism Varieties (Nos: 2008ZX0810-003 &2009ZX08001-022B), the post-doctoral foundation of China (No.20090450477).
References
Altpeter F., Baisakh N., Beachy R., Bock R., Capell T., Christou P., Daniell H., Datta K., Datta S.J., Dix P., Fauquet C., Huang N., Kohli A., Mooibroek H., Nicholson L., Nguyen T.T., Nugent G., Raemakers K., Romano A., Somers D., Stoger E., Taylor N., and Visser R., 2005, Particle bombardment and the genetic enhancement of crops: myths and realities, Mol. Breed., 15(3): 305-327 doi:10.1007/s11032-004-8001-y
Chen H., Tang W., Xu C.G., Li X.H., and Zhang Q., 2005, Transgenic indica rice plants harbouring a synthetic cry2A* gene of Bacillus thuringiensis exhibit enhanced resistance against rice lepidopteran pests, Thero. Appl. Genet., 111(7): 1330-1337 doi:10.1007/s00122-005-0062-8 PMid:16187120
Fojtová M., Bleys A., BedÅ™ichová J., Houdt H.V., KÅ™ížová K., Depicker A., and KovaÅ™ík A., 2006, The transsilencing capacity of invertedly repeated transgenes dependends on their epigenetic state in tobacco, Nucleic Acids Res., 34(8): 2280-2293 doi:10.1093/nar/gkl180 PMid:16670434 PMCid:1456325
Horvath H., Jensen L.G., Wong Q.T., Kohl E., Ullrich S.E., Cochran J., Kannangara C.G., and Von Wettstein D., 2001, Stability of transgene expression, field performance and recombination breeding of transformed barley lines, Thero Appl Genet, 102(1): 1-11 doi:10.1007/s001220051612
Hua Z.H., Zhu X.F., Wu M.G., Yu Y.C., Zhao Y., Gao Z.Y., Yan M.X., and Qian Q., 2003, Inheritance of exo-genes integrated into the rice genomes, Acta Agronomica Sinica, 29(1): 44-48
Huang D.N, Li J.Y., Zhang S.Q., Xue R., Yang W., Hua Z.H., Xie X.B., and Wang X.L., 1998, New technology to examine and improve the purity of hybrid rice with herbicide resistant gene, Chin. Sci. Bull., 43(9): 784-787 doi:10.1007/BF02898961
Huang D.N., Zhu B., Yang W., Xue R., Xiao H., Tian W.Z., Li L.C., and Dai S.H., 1996, Introduction of cecropin B gene into rice (Oryza sativa L) by particle gun bombardment and analysis of transgenic plants, Science in China (Series C), 39(6): 652-661
Iyer L.M., Kumpatla S.P., Chandrasekharan M.B., and Hall T.C., 2000, Transgene silence in monocots, Plant Mol. Biol., 43(2-3): 323-346 doi:10.1023/A:1006412318311 PMid:10999414
Khush G.S., 2005, What it will take to feed 5.0 billion rice consumers in 2030, Plant Mol. Biol., 59(1): 1-6 doi:10.1007/s11103-005-2159-5 PMid:16217597
Kohli A., Griffiths S., and Palacios N., 1999, Molecular characterization of transforming plasmid rearrangements in transgenic rice reveals a recombination hotspot in the CaMV35S promoter and confirms the predominance of microhomology mediated recombination, Plant J., 17(6): 591-601 doi:10.1046/j.1365-313X.1999.00399.x PMid:10230059
Kohli A., Twyman R.M., Abranches A., Wegel E., Stager E., Christou P., and Stoger E., 2003, Transgene integration, organization and interaction in plants, Plant Mol. Biol., 52(2): 247-258 doi:10.1023/A:1023941407376 PMid:12856933
Kumpatla S.P., Teng W., Buchholz W.G., and Hall T.C., 1997, Epigenetic transcriptional silencing and 5-azacytidine-mediated reaction of a complex transgene in rice, Plant Physiol., 115: 361-373 doi:10.1104/pp.115.2.361
PMid:9342860 PMCid:158494
Lindsay H., Adams R.L., 1996, Spreading of methylation along DNA, Biochem. J., 320(Pt2): 473-478 PMid:8973555 PMCid:1217954
Lu Y.J., and Zheng K.L., 1992, A simple method for isolation of rice DNA, Chinese J. Rice Sci., 1: 47-48 (in Chinese with an English abstract)
Matzke A.J.M., and Matzke M.A., 1998, Position effect and epigenetic silencing of plant transgenes, Curr. Opin. Plant Biol., 1(2): 142-148 doi:10.1016/S1369-5266(98)80016-2
Meyer P., 1995, Understanding and controlling transgene expression, Trends Biotechnol., 13(9): 332-337 doi:10.1016/S0167-7799(00)88977-5
Peng S., Laza R.C., Visperas R.M., Sanico A.L., Gassman K.G., and Khush G.S., 2000, Grain yield of rice cultivars and lines developed in Philippines since 1996, Crop Sci., 40(2): 307-314 doi:10.2135/cropsci2000.402307x
Popelka J.C., and Altpeter P., 2003, Evaluation of rye (Secale cereale L.) inbred lines and their crosses f or tissue culture response and stable genetic transformation of homozygous rye inbred line L22 by biolistic gene transfer, Thero. Appl. Genet., 107(4): 583-590 doi:10.1007/s00122-003-1314-0 PMid:12830384
Register J.C. 3rd, Peterson J., Bell P.J., Bullock W.P., Evans I.J., Frame B., Greenland A.J., Higgs N.S., Jepson I., Jiao S., Lewnau C.J., Sillick J.M., and Wilson H.M., 1994, Structure and function of selectable and non-selected transgenes in maize after introduction by particle bombardment, Plant Mol. Biol., 25(6): 951-961 doi:10.1007/BF00014669 PMid:7919215
Sambrook J., Fritsch E.F., and Maniatis T., 1989, Molecular Cloning: A Laboratory Manual, 3nd ed., Cold Spring Harbor Laboratory Press, Plainview, NY, pp.9.31-9.62
Tang W., Chen H., Xu C., Li X.H., Lin Y., and Zhang Q., 2006, Development of insect-resistant transgenic indica rice with a synthetic cry1C* gene; Mol. Breed., 18(1): 1-8 doi:10.1007/s11032-006-9002-9
Tang W., and Lin Y.J., 2007, Field experiment of transgenic crylAb insect resistant rice, HERIDITA (Bingjing), 29(8): 1008-1012 (in Chinese with an English abstract) doi:10.1360/yc-007-1008 PMid:17681932
Vain P., James V.A., Worland B., and Snape J.W., 2002, Transgene behaviour across two generations in a large random population of transgenic rice plants produced by particle bombardment, Thero. Appl. Genet., 105(6-7): 878-889 doi:10.1007/s00122-002-1039-5 PMid:12582913
Wang W., and Lin Y.J., 2007, Field experiment of transgenic cry1Ab insect resistant rice. Yi Chuan (Heriditas), 8: 1008-1012 (in Chinese with an English abstract)
Wu J.H., and Luo R.L., 2007, Current situation and strategy on international development of Chinese hybrid rice, Chinese Rice, 1: 11-13, 62 (in Chinese with an English abstract)
Zhang Y., Yin X., Yang A., Li G., Zhang J., 2005, Stability of inheritance of transgenes in maize (Zea mays L.) lines produced using different transformation methods, Euphytica, 144(1-2): 11-22 doi:10.1007/s10681-005-4560-1
. PDF(580KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Yan Zhao
. Longbiao Guo
. Huizhong Wang
. Danian Huang
Related articles
. Transgene
. Heredity and expression
. Hybriding transmission
. Particle bombardment
. Rice
Tools
. Email to a friend
. Post a comment


