2. Plant Protection and Quarantine Station of Hainan Province, Haikou, 570203, P.R. China
 Author
Author  Correspondence author
Correspondence author
Molecular Plant Breeding, 2011, Vol. 2, No. 12 doi: 10.5376/mpb.2011.02.0012
Received: 19 May, 2011 Accepted: 04 Jul., 2011 Published: 18 Jul., 2011
Liu et al., 2011, Genetic Diversity of Coconut Cultivars in China by Microsatellite (SSR) Markers, Molecular Plant Breeding Vol.2 No.12 (doi: 10.5376/mpb.2011.02.0012)
Assessment of genetic diversity is an essential component in germplasm characterization and utilization. In this study, we determined genetic diversity of 10 coconut (Cocos nucifera L.) accessions from six locations in Hainan province, China by using microsatellite markers. From the used 26 simple sequence repeat (SSR) markers, we detected a total of 188 alleles with an average of 7.23 alleles per locus and an average polymorphism information content of 0.575. Expected heterozygosity (He) of Haikou green Tall (HK-GT) was significantly higher than that of other Tall types, while the lowest heterozygosity was observed in Sanjiang green Tall (SJ-GT). At the genetic differentiation index (FST) of 0.078, they showed a low level of population differentiation. In addition to diversity parameters, Bayesian assignment tests and cluster analysis were used to determine population structure. Our study provided a better understanding of individual identities, genealogical relationships and geographical origin of coconut germplasm, and it could contribute to more efficient conservation and utilization of this germplasm.
The coconut palm (Cocos nucifera L., Arecaceae) is the most widely cultivated crop in Philippines, Indonesia, India, Sri Lanka and China, where coconut palm plays an important role in economy. It provides food supply and industrial products, such as coconut oil, copra, liquid endosperm and desiccated coconut. Almost every part of the coconut tree can be used in either making commercial products or meeting the food requirements of rural communities (Teulat et al., 2000). Coconut palms play an important role in the economy of Hainan province, directly providing food and income from coconut products. Moreover, it can be indirectly used as important features of the landscape, where tourism is a major component in the economy. The planting area of coconut is 43,300 hectares across Hainan Island as both economic and ornamental plants, and its yield is about 214 million coconuts (Guo, 2005, http://www.fao.org/docrep/010/ag117e/AG117E07.htm). However, coconut cultivation is confronted with a relative decline in many countries as the explosive competition from other oil crops, the increased demand of timber, drought, pest, disease and low fertility of the soils. Furthermore, the slow growth and long pre-breeding period of palm inhibit the genetic enhancement of coconut palm for productivity and tolerance to biotic and abiotic stresses (Rajesh et al., 2008). Germplasm collections, containing significant amount of genetic diversity within and among coconut populations, are essential for an effective crop improvement. Therefore, the assessment of genetic diversity within coconut populations becomes increasingly significant in germplasm conservation and utilization.
Investigation of coconut genetic diversity provides sufficient scientific data for germplasm management. Diversity analysis in coconut palm has been done by morphological traits, biochemical and molecular markers. Morphological and biochemical markers have shortages as follows: Long juvenile phase, high cost, long-term of field evaluation, environment factors and limited number of available phenotypic markers (Sugimura et al., 1997; Manimekalai et al., 2006). However, since molecular markers are detectable at all stages of development and can cover the entire genome, they, which detect variation at DNA level, overcome most limitations of morphological and biochemical markers, (Ashburner et al., 1997; Lebrun et al., 1998; Rohde et al., 1995; Perera et al., 1998, 1999, 2000, 2001, 2003; Rivera et al., 1999; Teulat et al., 2000; Dasanayake et al., 2003; Upadhyay et al., 2004; Zizumbo-Villarreal et al., 2006; Manimekalai et al., 2006, 2007). Among various available molecular marker techniques, simple sequence repeat (SSR) markers provide good signal in evaluating genetic diversity and genetic relationships in plants. The increased number of SSR markers greatly improves the previously established genetic relationships among coconut varieties/populations.
The first coconut type introduced to China from Southeast Asia is traditional Tall during Han Dynasty approximately 2000 years ago (Tang et al., 2006). In Hainan province, the palms are traditionally planted around villages, along roadsides and within cities, while the large plantations can be found in some locations. This cross-pollinating Tall coconut belongs to the Pacific group A3 (Lebrun et al., 2005). The self-pollinating Dwarf coconut was introduced from Malaysia during the 20th century (Martinez et al., 2009), belonging to the Pacific group A1 (Lebrun et al., 2005). Hybrids between the Malayan Yellow Dwarf (MYD) and the local Tall are planted in a few large plantations. However, little information is available on the genetic diversity among China coconut varieties/populations. The coconut is extensively planted in Hainan province (especially in Wenchang, Qionghai and Lingshui), where it is an integral part of spiritual and social life of indigene. For sustainable breeding, adoption and conservation in situ, it is necessary to develop a strategy to use diversity of coconut landraces for socio-economic benefits (Batugal and Oliver, 2003). In this study, we analyzed 45 coconut individuals from 10 accessions by using 30 SSR markers. Moreover, we had several objectives as follows: (i) to evaluate the genetic diversity of coconut varieties/populations in Hainan, China; (ii) to understand the genetic basis of chinese coconut varieties; (iii) to provide the groundwork for their genetic improvement and breeding; and (iv) to make good suggestions on protecting coconut varieties/populations in China.
1 Results and Analysis
1.1 Overall diversity parameters
In this study, we analyzed 45 individuals using 30 SSR primers. A total of 26 SSR primers showed greater allelic variability, which were clear, specific and reproducible. These 26 SSR primers revealed 188 alleles in the 45 individuals, whereas 163 alleles were polymorphic. The percentage of polymorphic alleles (PPA) was 84.65%. Table 1 shows that the polymorphic information content (PIC), observed heterozygosity (Ho), expected heterozygosity (He) of CAC03, CAC06, CAC08, CAC10, CAC21, CAC39 and CN11A10 exhibited higher levels than those of other loci, indicating that these seven primers are suitable for detecting the genetic diversity of coconut accessions in China. Furthermore, SSR analysis also showed that the gene flow of coconut in Hainan was 0.279 5, suggesting that the gene exchange between individuals was limited. The estimated out crossing rate (t) was 1.594 7, suggesting that this species is a typical plant with a cross-pollinate system.
.png) Table 1 Related parameters for 26 SSR loci |
Table 1 and Table 2 show that the observed and expected heterozygosities were slightly higher than the average values of the Pacific group A3 (0.248 and 0.263, respectively), but they were lower than the average values of the Pacific group A4 (0.487 and 0.512, respectively). The overall genetic structure in coconuts from Hainan province was explored by Wright's F statistics (Table 3). The FIT expresses the deviation of the whole Tall population from Hardy-Weinberg equilibrium. It was significantly different from zero, showing a slight deficit of heterozygotes. The FST represents the contribution of the divergence between populations to this deficit, which was also positive and significantly different from zero, indicating that allelic frequencies are different among accessions. If the whole population is in the Hardy-Weinberg equilibrium, the FST should be 0.078, which corresponds to the upper value of the confidence interval and suggests no differentiation among plantations.
.png) Table 2 Average numbers of alleles amplified and observed (Ho) and expected (He) heterozygosities for each accession |
Table 3 F statistics in the local Tall accessions |
The FIS corresponds to the within-population allelic correlations, which is influenced by inbreeding. The overall value was significantly different from zero, suggesting a slight tendency toward inbreeding. Slight but significant deviations from Hardy-Weinberg equilibrium indicate the differentiation among accessions as well as variation in the inbreeding level. However, this differentiation could be resulted from the genetic difference among populations or the fact that each accession is supposed to descend from a single tree.
1.2 Bayesian assignment tests among accessions
When we applied the Geneclass 2 assignment method with the local accessions as the reference dataset, the individuals of the hybrid accession from Wenchang were randomly assigned to one of them. Hybrids formed a distinct and homogenous group. Among the 42 local Tall, 19 of them were assigned to their own accessions, showing that there are some differences between accessions. The expected value for the null hypothesis of no differences between accessions was 5.7 (χ2=208***).
In spite of the deviations from the sampling protocol, members from the same accession still showed closer relationships than those from the different accessions. Three plantations (Wenchang, Changpo and Qionghai) were represented by two accessions with a total of 26 individuals (except WY78F1). Among them, 11 are assigned to their own accessions. Among the other remaining 15 individuals, four were assigned to the other accession of the same plantation. This result tends to weak the pattern suggested by the overall FST value (see above): A small part of the differences among accessions was due to the differences among the plantations.
1.3 Cluster analysis
The dendrogram was constructed from the genetic distance matrix by NTSYS program (Rohlf, 2000) (Figure 1). The 45 coconut individuals of HNT were divided into two main groups (i.e., Group â… and Group â…¡). Furthermore, Group â… was subdivided into two subgroups. This dendrogram shows that the members of the same plantation were not assigned to the same group as expected: E.g., both groups included Qionghai Green Tall (QH-GT), Xinglong Green Tall (XL-GT) and Wengchang Yellow Tall (WC-YT). Some accessions from different plantations were put into one group (e.g., CP-GT (Changpo Green Tall) 1, 5, 7 and QH-GT (Qionghai Green Tall) 1, 2, 3). This result is consistent with that from the Bayesin assignment tests.
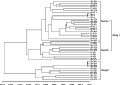 Figure 1 The dendrogram based on Cavalli-Sforza's distance showing genetic relationships among the 45 individuals of coconut in Hainan (China) |
2 Discussion
Genetic diversity is necessary to sustain the productivity of a crop since it furnishes new genes for yield, adaptation, disease resistance, high-value uses and characters (Frankel and Soulé, 1981). Rich diversity of coconuts exists in the field of farmers, and it is tremendously necessary to use this diversity to formulate strategies, thereby solving the problems of coconut farmers. In the present study, we characterized 10 accessions of coconuts by using the molecular tools in six coconut plantations in Hainan province. Moreover, we quantified the extent of genetic diversity and determined the population structure and its relationship with other coconut populations. Furthermore, our results also showed the relatively high genetic diversity of coconuts in Hainan province.
Our study has practical implications in farmer participatory evaluation and conservation of coconut genetic resources. Coconut accessions, which are adapted to the local conditions, high-yielding and possessing valuable characteristics, are under a threat of genetic erosion. In order to maintain coconut genetic resources, it is necessary to evaluate the extent of genetic diversity in the accessions. This process would make a significant contribution in promoting conservation of coconut germplasm in farmer′s fields through in situ and on-farm conservation (Batugal and Oliver, 2003). A better understanding of the diversity of coconuts that are available in farmers′ gardens would also help to utilize the range of germplasm for the sustainable production of coconuts, to increase the income of farmers and to make conservation strategies. Selected palms from these communities will be used as sources of seed nuts for planting, and community- managed seedling nurseries will be raised. This should serve as effective mechanisms for the introduction and promotion of farmer-preferred coconut diversity that could effectively support sustainable coconut production.
Meanwhile, our results showed that it is a powerful tool to address issues in individual assignment, population assignment and sibship reconstruction by combining the multi-locus SSR fingerprinting data with the model-based statistical method for individual assignment. Our study significantly improves the understanding of the earlier coconut collection from the Hainan province by clarifying and adding the passport data, which were largely missing in the records. In addition, the information on population structure, genealogical relationship and genotype identity in this germplasm group will be submitted to the International Coconut Genetic Resources Network (COGENT). This information will improve the efficiency and accuracy of coconut germplasm conservation, since inadequate representation of genetic diversity in the germplasm collections is another major constraint for efficiently conserving the genetic diversity of coconuts. The wild germplasm in the existing collections is primarily acquired during a few collecting expeditions, with the majority of them obtained in plain. In our study, a lot of wild germplasm were collected from tropical rainforest. Therefore, a fraction of this collection was probably cultivated varieties or derived from cultivated varieties. More collections are necessary to fill diversity gaps in the future.
This pattern suggests that cultivars from the same or different groups are equally important for coconut breeders in the search for valuable traits. The main objective in any plant genetic resources conservation program is to maintain the highest possible level of genetic variability covering across the gene pool of a given species or crop, both in its natural range and in a germplasm collection (Li et al., 2009). In order to preserve this valuable natural germplasm resource, we should make more efforts to protect its natural habitats as well as to sample an appropriate number of seeds from all these populations, which are either stored in a gene bank (e.g. seed bank, germplasm bank) or maintained as artificial populations in appropriate sites.
3 Materials and methods
3.1 Plant materials
In order to represent the overall genetic diversity of coconuts in Hainan, coconuts were collected from six different locations. The collection sites are either commercial or domestic plantations, locating in various parts of Hainan province (Table 4 and Figure 2). Tall coconuts are planted in all sites, whereas hybrids (WY, a hybrid between the local Tall and imported MYD) are planted in Wenchang.
.png) Table 4 Details of the accessions used in the study |
 Figure 2 Location map of the coconuts sampled in Hainan, China. The six different locations, where coconuts were collected, are indicated by red stars |
3.2 DNA isolation
The total DNA was isolated from 1 g fresh leaf using the CTAB method (Hoisington, 1992). Subsequently, the purified total DNA was quantified by gel electrophoresis, the DNA quality was verified by spectrophotometry, and DNA samples were stored at -20℃.
3.3 SSR analysis
A total of 30 pairs of highly polymorphic SSR primers in the microsatellite kit developed by Perera et al. (1999, 2000), Teulat et al. (2000) and Meerow et al. (2003) were used in this study. Table 5 lists the sequences of the primers. A 20-μL mixture was prepared for the PCR assay, containing 50 ng of template DNA, 200 μmol/L dNTPs, 1 U Taq DNA polymerse, 0.3 μmol/L forward SSR primer, 0.3 μmol/L reverse SSR primer, 50 mmol/L KCl, 10 mmol/L Tris-HCl (pH 8.3) and 2.5 mmol/L MgCl2. Briefly, the PCR reaction was performed at 94℃ for 5 min, then the amplification was carried out with 32 cycles at a melting temperature of 94℃ for 1 min, an annealing temperature of 55℃ for 90 sec, and an extension temperature of 72℃ for 2 min, followed by an additional extension at 72℃ for 5 min. Subsequently, the PCR products were analyzed on 8% polyacrylam- ide non-denaturing gels, and the bands were revealed by silver staining following the electrophoresis (Huo et al., 2005). The amplifications were done in duplicate for each analyzed primer.
.png) Table 5 List of coconut-specific microsatellite primer pairs with their sequences |
3.4 Diversity parameters
The amplified SSR markers were scored as present (1) or absent (0), and then recorded into a binary matrix as discrete variables (Lynch, 1994). The loci were eliminated from the analysis if they were not consistently amplified or with doubtful interpretation. We evaluated the genetic polymorphisms for different geographical regions of coconuts by calculating the number of alleles per locus (N), polymorphic loci (Np), percentage of polymorphic loci (P), expected heterozygosity (He), heterozygosity statistics for all loci (Ho), and Wright F statistics (Fis, Fst, Fit) (Nei, 1978). Based on the average expected and observed heterozygosities, the apparent outcrossing rate (t) was calculated from the inbreeding coefficient f (Ellstrand et al., 1978). All parameters of genetic diversity were calculated by the PopGene program (version 1.31).
3.5 Bayesian assignment tests
F statistics provided an overall assessment of differentiation among accessions. F statistics was examined more precisely by using a Bayesian assignment test. Individuals were be randomly assigned to any accession once no differentiation was observed. Therefore, deviation from randomness was the evidence of differentiation in one or more accessions. Geneclass 2 (Piry et al., 2004) was used, and 10 studied samples were used as the reference data. All individuals were tested and assigned to the most probable accession. In order to avoid systematic bias resulting from rare alleles, the “leave one out” option of Geneclass 2 was used: the tested individual was excluded from the reference dataset. Moreover, Bayesian assignment could also be used to identify populations demonstrating affinities with the local genotypes. In fact, the likelihood L of a population is the probability of obtaining the tested genotype in the population. Likewise, the probability of obtaining all Hainan Tall (HNT) genotypes is the product of the likelihoods. We used the sum of the scores issued by Geneclass 2 (-log L) as a dissimilarity measure.
3.6 Cluster analysis
A total of 45 coconut plants from Hainan (China) were studied via cluster analysis to determine their relationships. Cavalli-Sforza's distance rather than Nei's distance was used in this study because the role of genetic drift, migration and selection is more important than mutation in the evolution of this group. The weighted pair group method with arithmetic mean dendrogram was produced by the DARwin software (version 5.0.158) and the NTSYS software (version 2.1).
Authors’ contributions
XLL conceived the overall study, performed the experiment designs, and drafted the manuscript. HT and LHH took part to the data analysis and the writing. XLL obtained and analyzed the DDL data and was involved in the writing. All authors read and approved the final manuscript.
Acknowledgements
This research was supported by the National Natural Science Foundation (No. 30560092 and 31060259) and National Nonprofit Institute Research Grant of CATAS-ITBB. The authors thank the coconut farmers from the six cities for their assistance.
References
Ashburner G.R., Thompson W.K., and Halloran G.M., 1997, RAPD analysis of South Pacific coconut palm populations, Crop Science, 37(3): 992-997 doi:10.2135/cropsci1997.0011183X003700030048x
Batugal P., and Oliver J.T., 2003, Poverty reduction in coconut growing communities, vol.I: The Framework and Project Plan, International Plant Genetic Resources Institute Regional Office for Asia, the Pacific and Oceania (IPGRI-APO), Serdang, Selangor, Malaysia, pp.1-350
Dasanayake P.N., Everard E.H., Karunanayake H.G., and Nandadasa H.G., 2003, Characterization of coconut germplasm by microsatellite markers, Trop Agriculture Research, 15: 51-60.
Ellstrand N.C., Torres A.M., and Levin D.A., 1978, Density and the rate of apparent outcrossing in Helianthus annuus (Asteraceae), Systematic Botany, 3(4): 403-407 doi:10.2307/2418752
Frankel O.H., and Soulé M.E., 1981, Conservation and Evolution. Cambridge University Press, Cambridge, United Kingdom, pp.327
Hoisington D.A., ed., 1992, Laboratory Protocols, CIMMYT Applied Molecular Genetics Laboratory, Mexico DF, pp.1-86
Huo J.L., Zhang J., Luo G.Y., Pan W.R., Zhang M., and Zeng Y.Z., 2005, Sliver staining of microsatellite DNA-PAGE and simple method of making dry Gel, Progress In Veterinary Medicine, 26(1): 78-80
Lebrun P., Berger S.A., Hodgkin T., and Baudouin L., 2005, Biochemical and molecular methods for characterizing coconut diversity, Coconut genetic resources, In: Batugal P.A., Ramanatha Rao V., Oliver J. eds., Coconut genetic resources, International Plant Genetic Resources Institute-Regional Office for Asia, the Pacific and Oceania (IPGRI-APO), Serdang, pp.225-251
Lebrun P., N′Cho Y.P., Seguin M., Grivet L., and Baudouin L., 1998, Genetic diversity in coconut (Cocos nucifera L.) revealed by restriction fragment length polymorphism (RFLP) markers, Euphytica, 101(1): 103-108 doi:10.1023/A:1018323721803
Li M.M., Cai Y.L., Qian Z.Q., and Zhao G.F., 2009, Genetic diversity and differentiation in Chinese sour cherry Prunus pseudocerasus Lindl., and its implications for Conservation, Genetic Resources and Crop Evolution, 56(4): 455-464. doi:10.1007/s10722-008-9378-y
Lynch M., and Milligan B.G., 1994, Analysis of population genetic structure with RAPD markers, Molecular Ecology, 3(2): 91-99 doi:10.1111/j.1365-294X.1994.tb00109.x PMid:8019690
Manimekalai R., and Nagarajan P., 2006, Assessing genetic relationships among coconut (Cocos nucifera L.) accessions using inter simple sequence repeat markers, Scientia Horticulturae, 108(1): 49-54 doi:10.1016/j.scienta.2006.01.006
Manimekalai R., and Nagarajan P., 2007, Use of Simple Sequence Repeat markers for estimation of genetic diversity in coconut (Cocos nucifera L.) germplasm accessions, Journal of Plant Biochemistry and Biotechnology, 16(1): 29-33
Martinez R.T., Baudouin L., Berger A., and Dollet M., 2009, Characterization of the genetic diversity of the Tall coconut (Cocos nucifera L.) in the Dominican Republic using microsatellite (SSR) markers, Tree Genetics & Genomes, 6(1): 73-81 doi:10.1007/s11295-009-0229-6
Meerow A.W., Wisser R.J., Brown J.S., Kuhn D.N., Schnell R.J., and Broschat T.K., 2003, Analysis of genetic diversity and population structure within Florida coconut (Cocos nucifera L.) using microsatellite DNA, with special emphasis on the Fiji Dwarf cultivar, Theoretical and Applied Genetics, 106(4): 715-726
PMid:12596002
Nei M., 1978, Estimation of average heterozygosity and genetic distance from a small number of individuals, Genetics, 89(3): 583-590
PMid:17248844 PMCid:1213855
Perera L., Russell J.R., Provan J., McNicol J.W., and Powell W., 1998, Evaluating genetic relationships between indigenous coconut (Cocos nucifera L.) accessions from Sri Lanka by means of AFLP profiling, Theoretical and Applied Genetics, 96(3-4): 545-550 doi:10.1007/s001220050772
Perera L., Russell J.R., Provan J., and Powell W., 1999, Identification and characterization of microsatellites in coconut (Cocos nucifera L.) and the analysis of coconut population in Sri Lanka, Molecular. Ecology, 8(2): 344-346 PMid:10065554
Perera L., Russell J.R., Provan J., and Powell W., 2000, Use of microsatellite DNA markers to investigate the level of genetic diversity and population genetic structure of coconut (Cocos nucifera L.), Genome, 43(1): 15-21
doi:10.1139/g99-079 PMid:10701108 doi:10.1139/gen-43-1-15 PMid:10701108
Perera L., Russell J.R., Provan J., and Powell W., 2001, Levels and distribution of genetic diversity of coconut (Cocos nucifera L., var. Typica form typica) from Sri Lanka assessed by microsatellite markers, Euphytica, 122(2): 381-389 doi:10.1023/A:1012987224319
Perera L., Russell J.R., Provan J., and Powell W., 2003, Studying genetic relationships among coconut varieties/populations using microsatellite markers, Euphytica, 132(1): 121-128 doi:10.1023/A:1024696303261
Piry S., Alapetite A., Cornuet J.M., Paetkau D., Baudouin L., and Estoup A., 2004, GENECLASS2: a software for genetic assignment and firstgeneration migrant detection, Journal of Heredity, 95(6): 536-539 doi:10.1093/jhered/esh074 PMid:15475402
Rajesh M.K., Arunachalam V., Nagarajan P., Lebrun P., Samsudeen K., and Thamban C., 2008, Genetic survey of 10 Indian coconut landraces by simple sequence repeats (SSRs), Scientia Horticulturae, 118(4): 282-287.
doi:10.1016/j.scienta.2008.06.017
Rivera R., Edwards K.J., Barker J.H.A., Arnold G.M., Ayad G., Hodgkin T., and Karp A., 1999, Isolation and characterization of polymorphic microsatellites in Cocos nucifera L, Genome, 42(4): 668-675 doi:10.1139/g98-170 PMid:10464790 doi:10.1139/gen-42-4-668 PMid:10464790
Rohde W., Kullaya A., Rosriquez J., and Ritter E., 1995, Genetic analysis of Cocos nucifera L. by PCR amplification of spacer sequences separating a subset of copia-like Eco RI repetitive elements, Journal of Genetica and Breed, 49: 179-186
Rohlf F.J., 2000, NTSYS/pc, Numerical Taxonomy and Multivariate Analysis System, Version 2.1. Exeter Publications, New York, NY
Sugimura Y., Itano M., Salud C.D., Otsuji K., and Yamaguchi H., 1997, Biometric analysis on diversity of coconut palm: cultivar classification by botanical and agronomical traits, Euphytica, 98(1-2): 29-35 doi:10.1023/A:1003053128120
Tang B.N., Tang M., Chen C.F., Qiu P.H., Liu Q., Wang M.Y., and Li C.E., 2006, Characteristics of soil fauna community in the Dongjiao coconut plantation ecosystem in Hainan, China, Acta Ecologica Sinica, 26(1): 26-32
doi:10.1016/S1872-2032(06)60003-6
Teulat B., Aldam C., Trehin R., Lebrun P., Barker J.H.A., Arnold G.M., Karp A., Baudouin L., and Rognon F., 2000, An analysis of genetic diversity in coconut (Cocos nucifera L.) populations from across the geographic range using sequence-tagged microsatellites (SSRs) and AFLPs, Theoretical and Applied Genetics, 100(5): 764-771
doi:10.1007/s001220051350
Upadhyay A., Jayadev K., Manimekalai R., and Parthasarathy V.A., 2004, Genetic relationship and diversity in Indian coconut accessions based on RAPD markers, Scientia Horticulturae, 99(3-4): 353-362 doi:10.1016/S0304-4238(03)00103-1
Zizumbo-Villarreal D., Ruiz-Rodriguez M., Harries H., and Colunga-Garcia M.P., 2006, Population genetics, lethal yellowing disease and relationships among Mexican and imported coconut ecotypes, Crop Science, 46(6): 2509-251 doi:10.2135/cropsci2005.12-0462
. PDF(272KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Xiaolei Liu
. Hua Tang
. Dongdong Li
. Liheng Hou
Related articles
. Coconut cultivar
. Simple sequence repeat (SSR)
. Genetic diversity
. Hainan
Tools
. Email to a friend
. Post a comment


