Development of new set of microsatellite markers in cultivated tobacco and their transferability in other Nicotiana spp. 
2. Biotechnology section, Crop Improvement Division, Indian Institute of Rice Research Institute (ICAR-IIRR), Hyderabad, India
3. National Research Centre on Plant Biotechnology (NRCPB), New Delhi, India
 Author
Author  Correspondence author
Correspondence author
Molecular Plant Breeding, 2015, Vol. 6, No. 16 doi: 10.5376/mpb.2015.06.0016
Received: 28 Jul., 2015 Accepted: 29 Aug., 2015 Published: 14 Oct., 2015
Madhav M.S., Siva Raju K., Gaikwad K., Vishalakshi B., Murthy T.G.K. Umakanth B., 2015, Development of new set of microsatellite markers in cultivated tobacco and their transferability in other Nicotiana spp., Molecular Plant Breeding, 6(16) 1-13 (doi: 10.5376/mpb.2015.06.0016)
Scarcity of molecular markers in tobacco has been a limitation, hampering the acceleration of breeding efforts. Development of microsatellite markers is a prerequisite for mapping, tagging of many useful qualitative and quantitative traits and also for the generation of saturated linkage map. Use of microsatellite-enriched genomic libraries an efficient and rapid method for the identification of clones harboring microsatellite motifs leading to the development of microsatellite markers. In the present study, a total of 111 microsatellite motifs was identified from the enriched library, of which, 70 motifs (which includes perfect and imperfect repeat) were used for marker development. These newly developed markers could successfully differentiated different types of tobacco and diverse cultivars of Flue Cured Virginia (FCV) tobacco. The high rate of transferability (95-7% - 100%) of these microsatellite markers in a wide range of Nicotiana species indicated their potential as viable resources in the inter-specific gene transfer programme. The set of microsatellite markers developed in this study is a valuable addition to the already available DNA marker resources in tobacco.
1 Background
Tobacco (Nicotiana tabacum L) is one of the major commercial crops, belonging to the family Solanaceae, which contains many genuses, including Nicotiana. Several efforts are being undertaken to improve this crop, which could not be accelerated due to the non-availability of sufficient number of molecular markers. It is necessary to develop adequate molecular markers for reducing the time, effort and cost in breeding programs like gene targeting, selection and pyramiding. These molecular genetic markers have become useful tools to provide a relatively unbiased estimation of genetic diversity in plants. PCR-based markers like randomly amplified polymorphic DNA (RAPDs), amplified fragment length polymorphism (AFLP) and simple sequence repeats (SSRs) have an apparent advantage as cultivar descriptors since they are unaffected by environmental or physiological factors (Bowditch et al., 1993; Khan et al., 2008). Among the different types of molecular markers, SSRs are more abundant, ubiquitous in presence, hyper variable in nature and exhibit high polymorphism (Gupta et al., 1996; Senan et al., 2014). Moreover, they are highly reproducible, easy to apply, less cost, less laborious and hence used for cultivar identification in many crop plants such as barley (Russel et al., 1997;Maniruzzaman et al., 2014), soybean (Song et al., 1999; Bisen et al., 2015), potato (Barandalla et al., 2006; Carputo et al., 2013), Solanum melongena (Behera et al., 2006), rice (Yadav et al., 2008; Molla et al., 2015) and jute (Khan et al., 2008). Microsatellites have been shown to be almost twice as informative as dominant markers (RAPD and AFLP) and much more informative than RFLPs in soybean (Powell et al., 1996) and approximately six times more informative than RAPD (Rajora and Rahman, 2003). In addition to their use in determining the genomic structure and organization, these markers are also useful for the development of improved variety through marker aided selection. Through some microsatellite markers were reported by few earlier studies in tobacco (Bindler et al., 2007; 2011;Tong et al., 2012; Hughes et al., 2014), their number is limited as compared to other commercial crops, thereby demanding the development of more markers.
Generally, tobacco cultivars are distinguished by morpho-physio-chemical characters, but this method is slow, unrealistic and identification based on morphological traits is prone to environmental variations (Nelson, 1985). At the same time, a low level of genetic diversity exists within and among cultivated tobacco types (Ren and Timko, 2001). Cultivars that are closely related or less diverse cannot be easily distinguished by morphological indices (Degani et al., 1998). Accurate identification of cultivars has become increasingly important in realm of Plant Breeders Rights and cultivar registration. Hence, the molecular marker-based diversity analysis among the cultivars will provide the precise data for cultivar identification. The frequency of microsatellites in plant genomes is low as compared to animal genomes (Maguire et al., 2000; Squirrell et al., 2003) and this feature makes the isolation of microsatellites, a technically demanding task. With an objective of developing more SSR markers, the present study helped in the identification of microsatellite motifs using microsatellite enriched library, developed 70 SSR markers and demonstrated their use in the understanding of genetic diversity among different types of tobacco. In addition to this, transferability of these markers across different species of Nicotiana was tested and a minimal set of markers that could differentiate various species of Nicotiana was identified.
2 Results
2.1 Identification of microsatellites loci from the tobacco enriched library
Screening of microsatellite-enriched library of N. tabacum cv. Jayasree resulted in the identification of 1152 putative recombinants, which were then sequenced. Good quality sequences were obtained for 946 clones and all sequences (100%) contained microsatellite motifs. However, only 224 sequences were considered for designing primers. Considering the 224 clones possessing class I and class II SSRs as described by Mc Couch et al., 2002, AG/TC repeats were found to be abundant (56 clones, 25.3%) followed by CT/GA (38 clones, 17.2%), AC/TG (25 clones, 11.3 %), GT/CA (20 clones, 9.0%) and rest of the clones contained AG and CT compound repeats. The repeat length was highly variable at these loci with maximum for TbM13 with (GA)29 and TbM22 with (AGA)24. These sequences were submitted to NCBI Genbank and details of the sequence with accession numbers were tabulated (Table 2).Seventy motifs containing class I SSRs were targeted for marker development, of which, thirty nine (55.8%) were perfect repeats [(TC/AG)n, (GT/CA)n, (GA/CT)n and (AC/TG)n,], five (7.1%) were compound repeats and twenty six (37.1%) were interrupted repeats. The homology searches with the primers of these markers against the primers of markers reported earlier by Bindler et al., 2011; Tong et al., 2012 showed no significant homology.
.jpg) Table 2 Primer sequences, accession numbers, and characteristics of the 70 SSR markers in 24 Nicotiana species |
2.2 Diversity among Flue Cured Virginia (FCV) tobacco
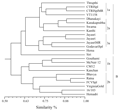
All the 70 SSR markers had shown clear and robust amplification with expected product size. Further, these markers were used in the diversity analysis of 24 FCV varieties of N. tabaccum. The pair-wise similarity measures among the tested varieties ranged from 0.54 to 0.91 revealing a broad genetic base (Figure 1). The varieties were mainly grouped into two main clusters. Cluster-A consisted of varieties developed indigenously by different breeding methods, while Cluster-B consisted of exotic introductions and their derivatives. The mean genetic similarity among the indigenously developed cultivars was 0.72.
2.3 Characterization of different types of tobacco:
The microsatellite marker analysis of different types of Indian tobaccos revealed that the varieties belonging to the same category were grouped together. There were different clusters for each tobacco type and the clustering pattern supports the traditional classification, except for the grouping of GC-2 (chewing type) and Dharla (Hookah type) in the same cluster (Figure 2). The genetic diversity analysis of 12 accessions from eight types of cultivated tobacco using 35 microsatellite markers revealed that all markers were polymorphic exhibiting an average polymorphic information content (PIC) of 0.55 (range 0.15-0.99) (Table 3). A total of 260 alleles were produced by these loci with an average of 7.4 alleles per marker in the size range of 150 to 500 bp. Among these markers, TbM28 had the highest PIC value of 0.99 followed by TbM9, TbM10, TbM14, TbM16 and TbM30 (0.97). It was observed that the mean PIC value was higher for markers developed by targeting perfect repeats (0.57), while it was lower for markers targeting imperfect repeats (0.19). The markers with higher PIC values will be useful for diversity analysis, varietal identification, mapping of traits, etc. The type specific markers identified in the present study would be useful in testing of purity of different types of tobaccos (Table 4).
.jpg) Table 4 List of markers for Nicotiana species identity and types/group identity within cultivated tobacco-Nicotiana tabacum |
2.4 Understanding the diversity among the genus Nicotiana
A total of 455 alleles was obtained from all 70 markers amplified using 24 Nicotiana spp. with an average of 6.4 alleles per locus (Table 2). A maximum of nine alleles was detected for four markers (TbM46, TbM52, TbM56 and TbM59) while a minimum of four alleles was detected for TbM4 with the allele size ranging from 150 to 700 bp.
All the 70 markers were showed high observed heterozygosity (Ho) than expected heterozygosity (He). The expected heterozygosity ranged from 0 to 0.38 (mean 0.18) while the observed heterozygosity ranged from 0 to 0.50 (mean 0.43) (Table 2). The markers developed from both perfect and imperfect SSR motifs exhibited slight difference in He (mean 0.19) and Ho (0.16) values. Similarly, the mean value of Ho for di and trinucleotide repeats were also showed very less variation, while the He values for di nucleotides (0.18) were slightly lesser than trinucleotides (0.21). The estimate for the frequency of null alleles was not zero for 19 of the 70 loci. The probabilities of identity ranged from 0.13 to 0.44 with a mean value of 0.13.
The genetic diversity analysis among the genus Nicotiana using Unweighted Pair Group Method on Arithmetic averages (UPGMA) method of clustering revealed that the Nicotiana species studied were separated into three main clusters (Figure 3). The cluster-1 is formed by the 11 species; all the species are belongs to the subgenus Rustica and Petunoides. In this cluster the species gluaca and paniculta formed small sub cluster (Ia), similarly, thrysiflora, rustica and undulate formed sub cluster (Ib). Among the cluster-1, rependa and nesophila were showed higher similarity (91%), since both are belong to the section Rependae and have the similar chromosome number 24. The second cluster formed with 7 species of Nicotiana, in which all the species are belongs to the subgenus Petunioides and section Suaveolentes except N. tabaccum which belongs to the subgenus Tabacum. The third cluster is formed by 6 species, among those 3 species belongs to the section Suaveolentes and rest are belongs to the different sections of subgenus Petunoides.
2.5 Transferability of tobacco SSRs to other Nicotiana species
Amplification of the 70 SSRs in 23 species of Nicotiana revealed an amplification of at least one allele and the cross transferability of these markers ranged from 95.7% to 100% (Table 5). All the markers developed in the present study were transferable to eight species and 26 markers showed 46 exclusive alleles. However, no exclusive alleles were identified for N. undulata, N. nesophila and N. debanyi (Table 4). In general, about 20 Nicotiana species could be easily discriminated using a minimal set of 11 markers (TbM31, TbM33, TbM41, TbM43, TbM44, TbM46, TbM47, TbM49, TbM52, TbM60, and TbM61).
3 Discussions
Microsatellite markers are considered as versatile tools for genetic analysis and breeding applications due to their abundance in the genome, high degree of variability and reduced time, effort and cost etc. However, the availability of microsatellite markers is limited in tobacco. Although many molecular markers were recently reported (Bindler et al., 2007; 2011, Tong et al., 2012; Hughes et al., 2014), they are not sufficient enough to use in inter specific hybridization programs and to develop a core genetic map like rice(Orjuela et al., 2010). Hence, there is an immense need to develop additional markers for the construction of dense genetic map, which would be the starting point for genetic mapping and eventual cloning of important genes from this crop. Therefore, we have employed a genomic enrichment method to capture the SSR motifs for the development of new microsatellite markers. Earlier, Sethy et al., 2006, developed 74 functional sequence-tagged microsatellite sites (STMS) primer pairs in chickpea by using genomic enrichment method. However, no attempt has been made to isolate microsatellite markers from the tobacco genome using the enrichment procedure, though this provides an attractive choice for targeted microsatellite development.
The enrichment protocol used in the present study has resulted in the identification of clones containing the SSR motifs with 82% success rate, which is significantly higher than any conventional methods exhibiting varying efficiency ranging from 0.045% to 12% (Zane et al., 2002). The genomic enrichment method for capturing SSRs is fast and convenient to handle as compared to the conventional method involving the genomic library preparation and sequencing. Using the enrichment method, high efficiency of microsatellite identification was also achieved in several crops (Gaitan-Solis et al., 2002; Riaz et al., 2004; Lowe et al., 2004) with varying success rates. By employing microsatellite-enrichment procedure, Marinoni et al., 2003, obtained 23% of clones containing SSR motif in chestnut, while Acquadro et al., 2005, obtained 85% in Cynara cardunculus. All the clones harboring SSR motif were made publicly available through GenBank submissions (Table 2).
More number of dinucleotide repeats (AG/TC) was observed than any other repeats, Which supported the fact that di-nucleotide repeats are the most frequently occurring microsatellites in plants (Lagercrantz et al., 1993; Thomas and Scott, 1993; Wang et al., 1994). However, a significant proportion (35.7%) of tri- and hexa-nucleotide repeat motifs were also observed, which could have resulted due to the chance cross hybridization of dinucleotide probes with other repeat region of the genome. Sequence analysis of 952 clones yielded 221 sequences with class I and II SSR motifs (McCouch et al., 2002). About 70 class I microsatellite loci were targeted for marker development.
All the identified SSR motifs showed more observed heterozygosity than expected heterozygosity. Both perfect and imperfect SSR motifs showed less variation of mean He and Ho values. Whereas the mean Ho for di- and tri-nucleotide repeats were also found to be slight difference. The results with more number of alleles amplified per locus (6.4) combined with the mean observed heterozygosity (Ho) values; suggest that considerable polymorphism is present at these microsatellite loci. The same results were represented in the earlier studies, reported in chickpea (Sethy et al., 2006). The highest heterozygosity detected at SSR loci, is potentially meaningful because high heterozygosity would indicate that the plant population likely has a substantial amount of adaptive genetic variation to escape the effects of a control agent that limits the development and maintenance of the plants (Allendorf and Luikart, 2007; Maranho et al., 2014), in contrast to plant populations showing a lower level of heterozygosity.
The occurrence of null alleles has been pointed out as a possible problem associated with the use of microsatellite markers (Callen et al., 1993), which might result in an individual being scored as homozygote instead of heterozygous and thus resulting in loss of information. Our results indicated that out of 70 markers, 19 markers showed less positive values for the frequency of null alleles, might be regarded as loci with a possible occurrence of null alleles (O’Reilly and Wright 1995). On the other hand, the negative value for the frequency of null alleles (r) observed in most loci might associate the homozygotes with negative characters that have been eliminated by humans in cultivated germplasm (Marinoni et al., 2003). In the present study most of the markers showed (51 out of 70 ) negative values for the frequency of null allele occurrence, it indicates that developed markers in the present study are potential tools for the differentiation of tobacco species without losing any information.
In the present study, the probability of identity was moderately high, which represents the power of each microsatellite to contribute to a unique genotype for each species (Dangl et al., 2005). Polymorphic Information Content (PIC) provides an estimate of discriminatory power of a marker to differentiate genotypes based on both the number of alleles expressed and their relative frequencies (Nagal et al., 2010). The average of PIC value was 0.57 for perfect repeats where as for imperfect repeats 0.19, indicates an isolation of highly polymorphic microsatellites in Nicotiana species. These polymorphic markers were should provide sufficient level of genetic diversity to investigate the fine scale population structure and evaluate the breeding strategy (Xu et al., 2009).
In the present study, we have used microsatellite markers for revealing polymorphism among the different Nicotiana species and also assessed for their cross-species transferability.
All the species of Nioctiana principally divided based on the subgenus to which they belong and the number of chromosomes. The species of subgenus Petunoides have 9 sections, mostly species belong to the same section, were grouped together. For instance, two species of Rependae section and two species of Alatae section were grouped in the same cluster (Ib) with higher percentage of similarity. Similarly 6 species of section Suaveolentes used in the present study were grouped in cluster II.
The clustering pattern of the tobacco species based on the nuclear SSR profile supports the traditional classification of genus Nicotiana with exception of grouping of N. rustica and N. undulate. Among the subgenus Petunioides, the section Undulatae and Trigonophyllae are considered to be close to the subgenera Rustica, so the species N. undulata was grouped along with the species belongs to the subgenus Rustica as these two species shares the same number (12) of chromosomes (Khan and Narayan, 2007). The microsatellite-based dendrogram was congruent with existing sectional representations based on morphological, chromosome and DNA information (Moon et al., 2008). In general, the transferability rates observed in this study were higher than those reported earlier (Moon et al., 2008), which could be due to high rates of conservation of primer binding sites. A minimal set comprising of 11 markers was constituted for the identification of most of the Nicotiana species. However, it should be pointed out that only one accession was used for each species, as the objective was to demonstrate the feasibility of these markers for studying genetic variation and relationships at species level and a detailed study involving more number of accessions for each species is warranted to validate these markers. Perfect clustering of genotypes belongs to the all the eight tobacco types were observed with these markers. Hence, the type specific markers identified in the present study for various tobacco types as well as various species specific markers belongs to Nicotiana would be useful in testing of purity of different types of tobaccos, studying the genetic variation among varieties, true hybrid identification, monitoring introgression of the target gene(s) and relationships between different tobacco types within cultivated tobacco using the cultivars representing a wide range of morphological diversity present within N. tabacum. The devloped markers alsoo exhibited high PIC values, suggesting their potential for genetic polymorphism studies in cultivated tobacco.
In conclusion, a set of 70 new microsatellite markers was developed in tobacco through microsatellite- enrichment method with high success rate, highlighting the efficiency of this method for the isolation of SSR motifs in tobacco as well as other crop species, for which genome sequence is not available. These markers were characterized in tobacco and in other Nicotiana species, thereby demonstrating the utility of these markers in genetic diversity assessment. The study also demonstrated the cross-species transferability of these markers to other Nicotiana spp resulting in the identification of species-specific markers. The microsatellite-enrichment technique used for the capturing of SSRs in tobacco and the markers developed in this study will be useful for fingerprinting, genetic diversity analysis, linkage map construction and genetic mapping studies in tobacco.
4 Material and Methods
Genomic DNA was extracted from fresh leaves of N. tabacum cv. Jayasree using CTAB method (Murray and Thompson, 1980). Around 25 µg genomic DNA was taken in a nebulizer and passed through inert Nitrogen gas to fractionate into 0.1-1 kb size fragments. The nebulized fragments were run on the gel and the desired size range of fragments was eluted. These fragments were polished to make blunt ends and adapters were ligated to them. Four biotinylated SSR probes [(TC)15, (GT)15, (GA)15, (AC)15] rich in other plant species, were hybridized with adapter-ligated fragments. Approximately 200 ηg of the fragments (100 bp-1000 bp) were added to a single reaction mixture containing 4.2 × SSC (Saline-Sodium Citrate, pH 7.0), 0.07 % SDS (sodium dodecyl sulfate), and 10 ρmol biotinylated probe. The mix was incubated at 95°C for 5 minutes and chilled quickly on ice for 2 minutes. It was then kept at proper annealing temperatures (depending on the melting temperature of each probe) for an hour to perform annealing [60°C for probe (GT)15, 37°C for (AT)15 and 57°C for (GA)15]. Meanwhile, Dynabeads M-280 Streptavidin (10 µg/µl) was prepared by gently shaking the vial to obtain a homogenous slurry. About 20 µl of the beads slurry was transferred to a 1.5 ml tube and washed four times with 300 µl of bead washing buffer (1x TE + 100 mM NaCl). The beads were re-suspended with 50 µl of the same buffer, added into the fragment-probe mix and incubated at room temperature for 30 minutes with constant gentle agitation. After immobilization, the supernatant was removed by applying a magnetic field to precipitate the beads, which were attached to the SSR containing fragments that hybridized to biotinylated probe. The bead-probe-fragment complex was washed three times each for 5 minutes with 400 µl non-stringency washing buffer (1 x TE + 1 M NaCl) at room temperature. The complex was further washed with 400 µl of stringency buffer (0.2 x SSC + 0.1% SDS) for three times each for 5 minutes at room temperature. After the final wash, the washing buffer was completely removed and 40 µl of sterile water was added, tapped gently and incubated at 95°C for 5 minutes. The eluted solution containing single strand SSR-enriched fragments was cloned into the pGEM-T easy vector, and the recombinants were transformed to DH 10 B E. coli electro-competent cell by electroporation.
4.1 Microsatellite identification and primer design
The enriched library was screened with pooling strategy by picking up individual clones from the agar plate and kept into 96-well format plates for further growth and subsequent isolation of plasmid DNA. The DNA of all clones was isolated by using 96-well format Qiagen DNA extraction kit (Qiagen, USA). The isolated DNA was pooled into row and column-wise and screened by PCR using anchored PCR. The positive pools were screened by demerging into individual samples. The plasmids were sequenced using the Big Dye Terminator reaction kit on the ABI 3700 prism automated DNA sequencer (Applied Biosystems, USA). The microsatellite motif in each sequence was identified using the software SSRIT and each sequence was compared against the local and NCBI sequence databases in order to identify the redundant clones. The unique sequences were retained and submitted to GenBank. Primers were designed based on the sequences flanking the microsatellite motifs using the PRIMER 3.0 software (Rozen and Skaletsky, 1998) using the following criteria: (1) Primer length 18-25 nucleotide with an optimal length of 20 nucleotides (2) primer Tm=50-600C (3) amplified PCR product size 100-400 bp, and (4) an optimal 40% GC content. The presence of structures such as hairpin or short repeat motifs was also considered while designing the primers. All primers, which flank the perfect repeats of the form (N1N2)X or (N1N2N3)X; imperfect repeats of the form N1N2N1N2N2N1N2N2N2N1N2; interrupted repeats of the form (N1N2)X(N)Y(N1N2)Z; and compound repeats of the form (N1N2)X(N3N4)Y were synthesized from IDT, Canada and they were standardized with gradient PCR to obtain the perfect annealing temperature to avoid non-specific amplification.
4.2 Testing of microsatellite markers
Each primer pair was tested by the amplification of genomic DNA isolated from the tobacco cultivar, Jayasree. PCR amplification was done in 25 µl reaction volume containing 200 mM Tris-HCl (pH 8.0), 500 mM KCl, 2 mM MgCl2, 0.25 mM of each dNTPs, 0.5 µM of each primer, 25 ng of genomic DNA and 1.0 U of Taq DNA polymerase (MBI Fermentas, Lithuania). Reactions were carried out in thermal cycler following the thermo-profile of an initial denaturation at 94ºC for 5 min followed by 35 cycles of 94ºC for 1 min, 55-57ºC for 1 min, 72ºC for 2 min and a final extension of 7 min at 72ºC. Amplified products were resolved on 6% polyacrylamide gels and stained with ethidium bromide. Initially, all the 70 primer pairs were used to amplify the DNA from 12 different tobacco types (Table 1) and one wild species of Nicotiana in order to establish their usefulness in the detection of intra- as well as inter-species-specific polymorphism. In order to check the transferability of these markers to other Nicotiana species, these primer pairs were further checked with 24 wild species of Nicotiana (Table 1).
4.3 Data analysis
The size of the alleles for each microsatellite locus was estimated by comparison with standard size DNA markers and scored across all the 24 Nioctiana species as well as genotypes from different types of cultivated tobacco using the gel documentation system (Alpha Innotech Corp., USA). A binary matrix was developed, in which 1 (one) represents the presence of an amplicon and 0 (zero) represents the absence of an amplicon. Genetic parameters like alleles per locus, the expected heterozygosity (He) were calculated as He = 1 – ∑ pi2 (Nei 1973), where pi represents the frequency of allele i among the varietal set. The observed heterozygosity (Ho) was obtained by direct calculation. The null allele frequency was calculated as r = (He – Ho)/(1 + He) (Brookfield, 1996). The probability of identity was calculated as PI = 1 – ∑pi4 + ∑∑ (2pipj)2 (Paetkau et al., 1995), where pi and pj represent the frequency of alleles i and j, respectively. Pair-wise genetic similarity was calculated among the 24 accessions of wild species as well as 12 genotypes belonging to different types of tobacco using Jaccard’s similarity coefficient. The similarity matrix was used for the construction of dendrogram using the UPGMA (Unweighted Pair Group Method with Arithmetic Averages) algorithm using the software, NTSYS-pc (version 2.1, Rohlf, 1998).
Authors Contribution
MS designed all experiments, carried out the major work and prepared Manuscript KS carried out the validation assays of markers KG developed microsatellite enriched library, BV participated in participated in the data analysis. TGK helped in Nicotiana species diversity analysis and BU: participated in data and statistical analysis. All authors read and approved the final manuscript.
Acknowledgements
MSM greatly acknowledge the support and financial assistance of Director, CTRI for carrying out the work.
References
Acquadro A., Portis E., and Lanteri S., 2003, Isolation of microsatellite loci in artichoke (Cynara cardunculus L. var. scolymus), Molecular Ecology Notes, 3(1): 37-39
http://dx.doi.org/10.1046/j.1471-8286.2003.00343.x
Barandalla L., Galarreta R., Rios D., and Ritter E., 2006, Molecular analysis of local potato cultivars from Tenerife Island using microsatellite markers, Euphytica, 152(2): 283-291
http://dx.doi.org/10.1007/s10681-006-9215-3
Behera T.K., Sharma P., Singh B.K., Kumar G., Kumar R., Mohapatra T., and Singh N.K., 2006, Assessment of genetic diversity and species relationships in eggplant (Solanum melongena L.) using STMS markers, Scientia Horticulturae, 107(4): 352-357
http://dx.doi.org/10.1016/j.scienta.2005.11.004
Bindler G., Hoeven V.R., Gunduz I., Plieske J., Ganal M., Rossi L., Gadani F., and Donini P., 2007, A microsatellite marker based linkage map of tobacco, Theoretical and Applied Genetics, 114(2): 341-349
http://dx.doi.org/10.1007/s00122-006-0437-5
Bindler G., Plieske J., Bakaher N., Gunduz I., Ivanov N., Hoeven R.V., Ganal M., and Donini P., 2011, A high density genetic map of tobacco (Nicotiana tabacum L.) obtained from large scale microsatellite marker development, Theoretical and Applied Genetics, 123(2): 219-230
Bisen A., Dhirendra Khare D., Priya Nair P., and Tripathi N., 2015, SSR analysis of 38 genotypes of soybean (Glycine Max (L.) Merr.) genetic diversity in India, Physiology and Molecular Biology of Plants, 21(1): 109-115
http://dx.doi.org/10.1007/s12298-014-0269-8
Bowditch B.M., Albright D.G., Williams J.G.K., and Braun M.J., 1993, Use of randomly polymorphic DNA in comparative genome studies, Methods in Enzymology, 224: 294-309
http://dx.doi.org/10.1016/0076-6879(93)24022-M
Brookfield J.F.Y., 1996, A simple new method for estimating null allele frequency from heterozygote deficiency, Molecular Ecology, 5(3): 453-455
http://dx.doi.org/10.1111/j.1365-294X.1996.tb00336.x
Callen D.F., Thompson A.D., Shen Y., Phillips H.A., Richards R.L, Mulley J.C., and Sutherland G.R., 1993, Incidence and origin of null alleles in the (AC) n microsatellite markers, American Journal of Human Genetics, 52(5): 922-927
Carputo D., Alioto D., Aversano R., Garramone R., Miraglia V., Villano C., and Frusciante L., 2014, Genetic diversity among potato species as revealed by phenotypic resistances and SSR markers. Plant Genetic Resource 21(1): 109-115
Degani C., Rowland L.J., Levi A., Hortynski J.A., and Gelletta G.J., 1998, DNA fingerprinting of strawberry (Fragaria xananassa) cultivars using randomly amplified polymorphic DNA (RAPD) markers, Euphytica, 102: 247-253
http://dx.doi.org/10.1023/A:1018385715007
Dorak T.V., 2014, www.dorak.info/genetics/popgen.html
Gaitan-Solis E., Duque M.C., Edwards K.J., and Tohme., 2002, Microsatellite repeats in common bean (Phaseolus
vulgaris), Crop Science, 42(6): 2128-2136
fighttp://dx.doi.org/10.2135/cropsci2002.2128
Gupta P.K., Balyan H.S., Sharma P.C., and Ramesh B., 1996, Microsatellites in plants: a new class of molecular markers, Current Science, 45(1): 45-54
Hughes P.W., Jaworski A.F., Davis C.S., Aitken S.M., and Simons A.M., 2014, Development of polymorphic microsatellite markers for Indian tobacco, Lobelia inflata (Campanulaceae), Applications in Plant Sciences, 2(4): 1300096
http://dx.doi.org/10.3732/apps.1300096
Khan H., Akter J., Islam M.S., Sajib A.A., Ashraf N., Haque S., and Khan H., 2008, Microsatellite markers for determining genetic identities and genetic diversity among jute cultivars, Australian Journal of crop science, 1(3): 97-107
Khan M.Q., and Narayan R.K.J., 2007, Phylogenetic diversity and relationships among species of genus Nicotiana using RAPDs analysis, African Journal of Biotechnology, 6(2): 148-162
Lagercrantz U., Ellegren H., and Andersson L., 1993, The abundance of various polymorphic microsatellite motifs differs between plants and vertebrates, Nucleic Acids Research, 21(5): 1111-1115
http://dx.doi.org/10.1093/nar/21.5.1111
Lowe A.J., Moule C., Trick M., and Edwards K.J., 2004, Efficient largescale development of microsatellites for marker and mapping applications in Brassica crop species, Theoretical for Applied Genetics, 108(6): 1103-1112
http://dx.doi.org/10.1007/s00122-003-1522-7
Maguire T.L., Edwards K.J., Saenger P., and Henry R., 2000, Characterization and analysis of microsatellite loci in a mangrove species, Avicennia marina (Forsk.) Vierh. (Avicenniaceae), Molecular Ecology, 9(11): 1853-1862
http://dx.doi.org/10.1046/j.1365-294x.2000.01089.x
Maniruzzaman., Talukder Z.A., Rohman S., Begum F., and Amiruzzaman M., 2014, Polymorphism study in Barley (Hordeum Vulgare) genotypes using microsatellite (SSR) markers, Bangladesh Journal of Agriculture Research, 39(1): 33-45
Marinoni D., Akkak A., Bounous G., Edwards K.J., and Botta R., 2003, Development and characterization of microsatellite markers in Castanea sativa (Mill.), Molecular Breeding, 11(2): 127-136
http://dx.doi.org/10.1023/A:1022456013692
McCouch S.R., Teytelman L., Xu Y., Lobos K.B., Clare K., Walton M., Fu B., Maghirang R., Li Z., Xing Y., Zhang Q., Kano I., Yano M., Fjellstrom R., De Clerk G., Schneider D., Cartinhour S., Ware D and Stein L., 2002, Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.), DNA Research, 9(6): 199-207
http://dx.doi.org/10.1093/dnares/9.6.199
Molla K.A., Debnath A.B., Ganie S.A., and Mondal T.K., 2008, Identification and analysis of novel salt responsive candidate gene based SSRs (cgSSRs) from rice (Oryza sativa L.), BMC plant Biology, 15: 122
http://dx.doi.org/10.1186/s12870-015-0498-1
Moon H.S., Nicholson J.S., and Lewis R.S., 2008, Use of transferable Nicotiana tabacum L. microsatellite markers for investigating genetic diversity in the genus Nicotiana, Genome, 51(8): 547-599
http://dx.doi.org/10.1139/G08-039
Murray M.G., and Thompson W.F., 1980, Rapid isolation of high-molecular- weight plant DNA, Nucleic Acids Research, 8(19): 4321-4325
http://dx.doi.org/10.1093/nar/8.19.4321
Nagl N, Taski-Ajdukovic K, Popovic A, Curcic A, Danojevic D, Kovacev L. Estimation of genetic variation among related sugar beet genotypes by using RAPD. Genetika. 2011; 43: 575–582, doi: 10.2298/GENSR1103575N
http://dx.doi.org/10.2298/GENSR1103575N
Orjuela J., Garavito A., Bouniol M., Arbelaez J.D., Moreno L., Kimball, Wilson G., Franc J., Rami O., Tohme J., McCouch S.R., and Lorieux M., 2010, A universal core genetic map for rice, Theoretical and Applied Genetics, 120(3): 563-572
http://dx.doi.org/10.1007/s00122-009-1176-1
Paetkau D., Calvert W., Stirling I., and Strobeck C., 1995, Microsatellite analysis of population structure in Canadian polar bears, Molecular Ecology, 4: 347-354
http://dx.doi.org/10.1111/j.1365-294X.1995.tb00227.x
Powell W., Morgante M., Andre C., Hanafey M., Vogel J., Tingey S., and Rafalski A., 1996, The comparision of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis, Molecular Breeding 2(3): 225-238
http://dx.doi.org/10.1007/BF00564200
Rajora O.P., and Rahman M.H., 2003, Microsatellite DNA and RAPD fingerprinting, identification and genetic relationships of hybrid poplar (Populus x canadensis) cultivars, Theoretical and Applied Genetics, 106(3): 470-477
Ren N., and Timko M.P., 2001, AFLP analysis of genetic polymorphism and evolutionary relationships among cultivated and wild Nicotiana species, Genome, 44(4): 559-571
http://dx.doi.org/10.1139/gen-44-4-559
Riaz S., Dangl G.S., Edwards K.J., and Meredith C.P., 2004, A microsatellite marker based framework linkage map of Vitis vinifera L. Theoretical and Applied Genetics, 108(5): 864-872
http://dx.doi.org/10.1007/s00122-003-1488-5
Rohlf F.J., 1998, NTSYSpc Numerical Taxonomy and Multivariate Analysis System Version 2.0 User Guide, Applied Biostatistics Inc., Setauket, New York, pp.37
Rozen S., and Skaletsky H.J., 1998, Primer 3, code available at http://www.genome_software/other/primer3.html
Russell J., Fuller J., Young G., Thomas B., Taramino G., Macaulay M., Waugh R., and Powell W., 1997, Discriminating between barley genotypes using microsatellites markers, Genome, 40(4): 442-450
http://dx.doi.org/10.1139/g97-059
Senan S., Kizhakayil D., Sasikumar B., Sheeja TE., 2014, Methods for Development of Microsatellite Markers: An Overview, Notulae Scientia Biologicae, 6(1): 1-13
http://dx.doi.org/10.15835/nsb.6.1.9199
Sethy N.K., Shokeen B., Keith J., Edwards., and Bhatia S., 2006, Development of microsatellite markers and analysis of intraspecific genetic variability in chickpea (Cicer arietinum L.), Theoretical and Applied Genetics, 112(8): 1416-1428
http://dx.doi.org/10.1007/s00122-006-0243-0
Song Q.J., Quigley C.V., Nelson R.L., Carter T.E., Boerma H.R., and Strachan J.R., 1999, A selected set of trinuclotide simple sequence repeat markers for soybean cultivar identification, Plant Varieties and Seeds, 12: 207-220
Squirrell J., Hollingsworth P.M., Woodhead M., Russell J., Lowe A.J., Gibby M., and Powell W., 2003, How much effort is required to isolate nuclear microsatellites from plants, Molecular Ecolology, 12(6): 1339-1348
http://dx.doi.org/10.1046/j.1365-294X.2003.01825.x
Thomas M.R., and Scott N.S., 1993, Microsatellite repeats in grapevine reveal DNA polymorphisms when analyzed as sequence-tagged sites (STSs), Theoretical and Applied Genetics, 86(8): 985-990
http://dx.doi.org/10.1007/BF00211051
Tong Z., Yang Z., Chen X., Jiao F., Li X., Wu X., Gao Y., Xiao B., and Wu W., 2012, Large-scale development of microsatellite markers in Nicotiana tabacum and construction of a genetic map of flue-cured tobacco, Plant Breeding, 131(5): 674-680
http://dx.doi.org/10.1111/j.1439-0523.2012.01984.x
Wang Z., Weber J.L., Zhong G., and Tanksley S.D., 1994, Survey of plant short tandem DNA repeats, Theoretical and Applied Genetics, 88(1): 1-6
http://dx.doi.org/10.1007/BF00222386
Waits L.P., Gordon Luikart G., and Taberlet P., 2001, Estimating the probability of identity among genotypes in natural populations: cautions and guidelines, Molecular Ecology, 10: 249-256
http://dx.doi.org/10.1046/j.1365-294X.2001.01185.x
Yadav S., Rana P., Saini N., Jain S., and Jain R., 2008, Assessment of genetic diversity among rice genotypes with different adaptations to salinity using physio-morphological and molecular markers, Journal of Plant Biochemistry and Biotechnology, 17(1): 1-8
http://dx.doi.org/10.1007/BF03263252
Zane L., Bargelloni L., and Patarnello T., 2002, Strategies for microsatellite isolation: a review, Molecular Ecology, 11(1): 1-16
http://dx.doi.org/10.1046/j.0962-1083.2001.01418.x
. PDF(677KB)
. FPDF
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Madhav M.S.
. Siva Raju K.
. Gaikwad K.
. Vishalakshi B.
. Murthy T.G.K.
. Umakanth B.
Related articles
. Tobacco
. Functionality
. Transferability
. Genetic diversity
. Microsatellite
Tools
. Email to a friend
. Post a comment

.jpg)
.png)
.jpg)
.jpg)


