Research Article
Analyses on Progenitor Donors of the Cultivated Allotetraploid Cottons Revealed by GISH 
2 Institute of Cotton Research of CAAS / State Key Laboratory of Cotton Biology, Anyang, Henan, 455000, China
 Author
Author  Correspondence author
Correspondence author
Molecular Plant Breeding, 2016, Vol. 7, No. 14 doi: 10.5376/mpb.2016.07.0014
Received: 12 Jan., 2016 Accepted: 27 Feb., 2016 Published: 04 Apr., 2016
Xiao S.P., Wang K.B., Yang L., Ke X.S., Wang C.Y., Liu X.W., Sun L.Q., Yang S.Q., Liu F., and Chen Y., 2016, Analyses on Progenitor Donors of the Cultivated Allotetraploid Cottons Revealed by GISH, Molecular Plant Breeding, 7(14): 1-10 (doi: 10.5376/mpb.2016.07.0014)
GISH (Genomic in situ hybridization) of the mitotic metaphase chromosomes of two cultivated tetraploid cotton (AD)1 (G. hirsutum) and (AD)2 (G. barbadense) with all 3 diploid A cotton gDNA(genomic DNA) as probes, blocking with ssDNA(salmon sperm DNA) respectively. The hybridization signals were dected distribute in the A sub-genome chromosomes of (AD)1 and (AD)2, besides, three pairs of crimson signals were also detected only with the A1-a gDNA probe. which were named GISH-NORs. GISH of (AD)1 and (AD)2 with all 13 diploid D cotton gDNA as probes, blocking with ssDNA respectively, except the D6 (G.gossypiodies) gDNA probe generated the hybridization signals in all the chromosomes of (AD)1 and (AD)2, the other 12 diploid D gDNA probes only generated the signals on the D sub-genome chromosomes of (AD)1 and (AD)2, the D6 gDNA probe was very specifical.And three pairs of strong GISH-NORs were detected with all 13 diploid D genome species gDNA probes, the intensity of their GISH-NORs were much brighter than the A1-a gDNA probe. These results visually confirmed the amphidiploid origin of the allotetraploid cotton species. DA (distinguishing ability ) values of each gDNA probe generated were calculated basing on the above GISH rsults.It showed that the DA value of A1-a gDNA probe was the biggest in all 3 diploid A genomes both in (AD)1 and (AD)2 GISH, and this indicated that A1-a genome was most likely to be the A sub-genome progenitor donor of (AD)1 and (AD)2, while the D3-d (G. davidsonii ) and D5 (G. raimondii) genome species were most likely to be the D sub-genome progenitor donor of (AD) 1, and (AD) 2 respectively. And this further confirmed that tetraploid cottons are polyphyletic.
Gossypium genus belongs to the Malvaceae family, which contains 46 diploid (2n = 2× = 26,) species and 5 tetraploid (2n = 4× = 52). The Gossypium genus are believed to have originated from a common ancestor approximately 5~10 million years ago,of which eight diploid genomes, designated as A to G and K, have been found across Africa, Asia Australia and North America (Phillips,1963; Wendel ,1989; Fryxell,1992; Dejooded and Wendel, 1992), and 5 tetraploid species were named (AD)1 (G. hirsutum), (AD)2 (G. barbadense), (AD)3 (G. tomentosum), (AD)4 (G. mustelinum) and (AD)5 (G. darwinii) respectively(Wendel ,1989). At present, (AD)1 and (AD)2 are major cultivated species, which are extensive cultivated in the world. Previous studies had shown that diploid cotton A and D genome species are the respective donor species of A and D sub-genome of the allotetraploid cotton species, and the A sub-genome chromosomes are longer than the D sub-genome chromosomes(Phillips,1966; Endrizzi,1985).
The Fluorescence in situ hybridization (FISH) technology was introduced to plant research in 1985 (Rayburn,1985), which has applied to various aspects such as repeat sequences positioning, chromosome identification, cell genetic map construction, the origin of polyploid genome evolution and phylogenetic relationships and so on (Snowdon et al., 1997 ; Tang et al., 2000; Jiang and Gill,2006; Wang k et al.,2008; Nemeth et al., 2015; Melo et al., 2015).
FISH technology have developed suceessfully on other crops (Mukai et al.,1993; Cheng et al., 2002 ; Xiong et al., 2004). But research on cotton far lags behind those, main because cotton cell has more density cytoplasm, more hard cell wall, more number of chromoses and smaller size compare with other crops, which lead to cotton cell processing more difficult in prepare of high quality chromosome slides. In recent years, with the breakthrough of the bottleneck technology, cotton FISH research is developing rapidly(Liu et al., 2005 ; Wang et al., 2009; Wu et al., 2008 ; Gan et al., 2013 ; Zhang et al., 2014; Cui et al., 2015).
At present, about the great theories on Gossypium such as origin of species, evolution and classification had already formed the basic agreement. However, there still exist dissensions on the specific donors of the 5 tetraploid cotton species, and the germplasm disproportionation in the process of origin, and the homologous degree between different genome species of Gossypium or within one genome species(Liu et al., 2003 ;Wang KB et al., 2008). Our laboratory has made a great progress on using FISH technology to research the origin and evolution of cotton genomes and the interspecific genetic relationship or the genonomy of Gossypium (Wang et al., 1999; Wang et al., 2001; Liu et al., 2005; Wu et al., 2008; Gan et al., 2013).
In this study, GISH(Genomic in situ hybridization, one of the FISH technology) was used to the two cultivated alltetraploid cottons somatic metaphase chromosomes of (AD)1 and (AD)2, and used All 3 diploid A and 13 diploid D cotton gDNA (genomic DNA) as probe blocking with ssDNA, in order to further explore the specific A and D sub-genome progenitor donors of (AD)1 and (AD)2, and their genetic relationships; homologous degree between each tested cotton genome species. Wished to provide a beneficial enlightenment and help for the genetic improvement and suggest the preferred Gossypium species for genome sequencing.
1 Results and Analysis
1.1 GISH of (AD)1 and (AD)2 with all 3 diploid A cotton gDNA as probes respctively
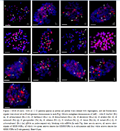
GISH of the somatic metaphase chromosomes of (AD)1and (AD)2 both with all 3 diploid A (A1, A2 and A1-a) gDNA as probes respectively, blocking with ssDNA(salmon sperm DNA).The results showed that the red hybridization signals were mainly distributed on the longer 13 pairs of A sub-genome chromosomes of (AD)1 and (AD)2 (As shown in Figure 1), and this visually confirmed the allodidiploid origin of the tetraploid cottons (Wendel et al., 2002). Besides, three pairs of crimson signals were detected only with the A1-a gDNA probe both in (AD) 1 and (AD) 2 (Figure 1-c, f ), which were named “GISH-NORs” (Liu et al., 2005) , of which one in the A sub-genome chromosomes (green arrows), and two in the D sub-genome chromosomes (white arrows), and it significantly different from the A1 and A2 gDNA probes, the specific distributions about GISH-NORs were shown in Table 1.
|
Figure 1 GISH of mitotic metaphase chromosomes of two cultivated allotetraploid cotton species G.hirsutum (AD) 1 and G.barbadense (AD) 2 with all 3 A diploid cotton gDNA probes respectively (all probes were labeled with digoxigenin, and red fluorescence signals were observed on A sub-genome chromosomes in each image). Figure 1-a To 1-c: GISH of G.hirsutum var. zhong 16 with diploid A genome species G.herbaceum var. hongxing (A1), G.arboreum var. shixiya 1 (A2), G.herbaceum wild species arfrium (A1-a) gDNA probe rsepectively, blocking with ssDNA (In Figure1-c, six arrows showed GISH-NORs, of which two green arrows denote the GISH-NORs in A sub-genome chromosomes and other four white arrows denote the GISH-NORs in D sub-genome chromosomes of (AD)1 ). Figure 1-d To 1-f: GISH of G.barbadense var. Xinhai 7 also with diploid A genome species A1, A2, and A1-a gDNA probe rsepectively, blocking with ssDNA (In Figure 1-f, six arrows also showed the signals of GISH-NOR, of which two green arrows denote the GISH-NORs in A sub-genome chromosomes and four white arrows denote in D sub-genome). Bars=10mm |
|
Table 1 Distribution of GISH-NORs in (AD) 1 and (AD) 2 by GISH with all A and D diploid gDNA probes |
1.2 GISH of (AD)1 and (AD)2 with all 13 diploid D cotton gDNA as probes respectively
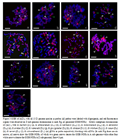
Except the D6(G.gossypiodies) gDNA probe, GISH of the somatic metaphase chromosomes of (AD)1 and (AD)2 both with other 12 diploid D gDNA as probe respectively, blocking with ssDNA (As shown in Figure 2 and Figure 3), the red fluorescence signals were observed on the short D sub-genome chromosomes of (AD)1 and (AD)2. In addition, three pairs of major GISH-NORs were detected, of which one in the longer A sub-genome chromosomes (green arrows showed in each image), and other two in the D sub-genome chromosomes (white arrows in each image),and the specific distribution about GISH-NORs were shown in Table 1. However, there were some differences on the fluorescence signal strength generated by each D genome species probe. GISH of (AD)1 and (AD)2 with D6 gDNA probe respectively, the red fluorescence signals were not only distribute on the D sub-genome chromosomes but also on A sub-genomes (As shown in Figure 2-h and Figure 3-h), and we could not distinguish the A and D sub-genome of (AD) 1 and (AD) 2, and three pairs of major GISH-NORs were also observed. This showed a great difference between the D6 genome and the other 12 D genomes, and the result illustrated that diploid D6 genome may contain plenty of A genome repeat sequences, it is a very special genome species in the diploid D genomes.
|
Figure 2 GISH of (AD) 1 with all 13 D genome species as probes (all probes were labeled with digoxigenin, and red fluorescence signals were observed on D sub-genome chromosomes in each Figure). Mitotic metaphase chromosomes of (AD) 1 with G. thurberi (D1) (a), G. armourianum (D2-1) (b), G. harknessii (D2-2) (c), G. klotzschianum (D3-k) (d), G. davidsonii (D3-d) (e), G. aridum (D4) (f), G. raimondii (D5) (g), G. gossypiodies (D6) (h), G. lobatum (D7) (i), G. trilobum (D8) (j), G. laxum (D9) (k), G. turneri,(D10) (l), G. schwendimanii (D11) (m) gDNA as probe respectively, blocking with ssDNA (In each Fig, there are six arrows, all arrows show signals of GISH-NORs, of which two green arrows denote the GISH-NORs in A sub-genome and four white arrows denote the GISH-NORs in D sub-genome). Bars=10mm. |
|
Figure 3 GISH of (AD) 2 with all 13 D genome species as probes (all probes were labeled with digoxigenin, and red fluorescence signals were observed on D sub-genome chromosomes in each Fig, all generated GISH-NORs). Mitotic metaphase chromosomes of (AD) 2 with G. thurberi (D1) (a), G. armourianum (D2-1) (b), G. harknessii (D2-2) (c), G. klotzschianum (D3-k) (d), G. davidsonii (D3-d) (e), G. aridum (D4) (f), G. raimondii (D5) (g), G. gossypiodies (D6) (h), G. lobatum (D7) (i), G. trilobum (D8) (j), G. laxum (D9) (k), G. turneri,(D10) (l), G. schwendimanii (D11) (m) gDNA as probe respectively, blocking with ssDNA (In each Fig, there are six arrows, all arrows show the GISH-NORs, of which two green arrows denote the GISH-NORs in A sub-genome while other four white arrows denote the GISH-NORs in D sub-genome). Bars=10mm. |
1.3 DA value analysis based on GISH of (AD)1 and (AD)2 with diploid A, D genome species probes
DA (distinguishing ability) value reflect the genetic relationship between the diploid A or D genomes and the tetraploid genomes (Markova et al., 2007). DA values (As shown in Table 2) generated by A1, A2 and A1-a gDNA probes to (AD)1 was 0.361, 0.358 and 0.369 respectively. It showed the A1-a gDNA possess the strongest ability to recognize the A sub-genome chromosomes of (AD) 1, while the A1 and A2 gDNA possess same ability to recognize the A sub-genome chromosomes of (AD) 1, that’s to say the A1-a genome had closer genetic relationship with the A sub-genome of (AD)1. And DA values generated by A1, A2 and A1-a gDNA probes to (AD)2 were 0.343, 0.346 and 0.352 respectively. the results also showed the A1-a gDNA possess the strongest ability to recognize the A sub-genome chromosomes of (AD)2, therefore, the A1-a genome also had closer genetic relationship with the A sub-genome of (AD)2.
|
Table 2 DA values of all diploid A, D genome species probes generated by GISH of (AD) 1 and (AD) 2 |
DA values of (AD)1 generated by all 13 diploid D gDNA probes respectively were shown in Table 2. Because of the hybridization signals generated by D6 gDNA probe were distributed on both A and D sub-genome chromosomes of (AD)1, therefore the DA value generated by D6 gDNA probe(DA value was 0.402) was much higher than the other 12 D gDNA probes in (AD)1. Except the D6 gDNA probe, the highest DA value was 0.286 generated by D3-d gDNA probe, and then followed by D4, D1, D5, D11, D2-1, D2-2, D8, D3-k, D9, D10 and D7 probes. This showed that the D3-d gDNA probe possess the strongest ability to recognize the D sub-genome chromosomes of (AD)1, while the other D gDNA probes show weaker ability to recognize the D sub-genome chromosomes of (AD) 1, therefore the D3-d genome had closer genetic relationship with the D sub-genome of (AD)1.
DA values of (AD) 2 generated by all 13diploid D gDNA probes also be shown in Table 2. Except the D6 gDNA probe(DA value was 0.387), the highest DA value generated by D5 gDNA probe was 0.263, and then followed by D3-d, D1, D4, D2-1, D2-2, D11, D8, D3-k, D10, D9 and D7 probes. which was also higher than those of the other 12 D gDNA probes. The results showed that the D5 gDNA possess the strongest ability to recognize the D sub-genome chromosomes of (AD)2, while the other 12 D gDNA show weaker ability to recognize the D sub-genome chromosomes of (AD)2, so the D5 genome species had closer genetic relationship with the D sub-genome of (AD)2.
2 Discussion
2.1 The A sub-genome progenitor of (AD)1 and (AD)2
Since the discovery that allotetraploid Gossypium genomes contain both A and D genomes, investigators had attempted to look for which one of the modern diploid A and D genome species can be best served as the progenitor genome donors of allopolyploid cottons.
Which one of the diploid A genome species was the really donor of the A sub-genome of allopolyploid cottons? Many previous researches suggested that G. herbaceum (A1) was the donor or the similar ancestors of the allopolyploid A sub-genome (Beasley, 1940; Gerstel, 1953; Phillips, 1963; 1964). However, the subsequent research had shown that there existed much differences between G. herbaceum (A1) and the A sub-genome of allopolyploid cotton, whether in the chromosome or molecular level (Wendel, 1989; Wendel and Albert, 1992ï¼›Cronn, et al., 1996). And cell cytogenetic and comparative mapping research also revealed that there existed at least two large translocations between their genomes (Gerstel, 1953ï¼›Small, et al., 1998ï¼›Liu and Wendel, 2001). Branch taxonomy analyses of the molecular sequences had showed that G. herbaceum(A1) was not the actual progenitor of the A sub-genome of allotetraploid cottons (Endrizzi, et al., 1985; Wendel and Cronn, 2002). In the evolutionary process of allotetraploid cottons, G. herbaceum(A1) and G. arboretum(A2) were the phylogenetically sisters between each other and hence were genealogical equidistant to the A sub-genome of the allotetraploid cottons (Cronn et al., 1996; Liu and Wendel, 2001; Wendel, 1989; Wendel and Albert, 1992).
In our experiment, GISH of (AD) 1 and (AD) 2 both with all 3 A genome gDNA as probes, 13 pairs of A sub-genome chromosomes were painted with red fluorescence signals, and the DA value was very similar between A1 and A2 gDNA probe, there was no significant difference between A1 and A2 both in GISH of (AD)1 and (AD)2, while the DA value of A1-a gDNA probe was higher than A1 and A2 gDNA probe, the A1-a gDNA had the strongest ability to recognize the A sub-genome chromosomes of (AD) 1 and (AD)2, so we considered that A1-a genome species was most likely to be the A sub-genome progenitor donor of allotetraploid cotton (AD)1 and (AD)2.
The A1-a genome had been divided into the variant of A1 genome in the taxonomy(Stewart, 1987). Many researches always used the varieties of A1 and A2 genome species to study the origination and evolution of the A sub-genome of allotetraploid cottons, but they seldom and nearly didn’t use the A1-a genome as the research materials, and even some researchers took the A1-a and A1 genome to lump together (Stewart, 1987; Wendel,1989; Wendel and Albert, 1992; Wendel et al.,1994, 1995; Wendel and Wessler, 2000; Wendel and Cronn, 2002; Wang et al., 2004). In this experiment, when using A1-a gDNA as probe we found that the GISH result was obviously distinguished from the A1 gDNA probe, and Wang et al (1995) had also found the karyotype parameters exist significant differences between A1 and A1-a genome. The A1-a genome made a strong independence from A1 genome, and we suggested that A1-a genome should be given the “species” level in the classification of gossypium, that’s means the A1-a genome possessed the same status with A1 and A2 genome.
2.2 The D sub-genome progenitor of (AD)1 and (AD)2
Since the discovery that allopolyploid Gossypium species contain two genomes whose progenitors presently occur in different hemispheres, investigators had attempted to provide pieces to the puzzle of the polyploid origin. A diverse array of tools had been used in an effort to examine this issue, from early study methods by comparative morphology, cytology, cytogenetic, comparative phytochemistry, and protein electrophoretic methods to modern phylogenetic investigations using DNA sequencing of homologous genes. Several investigators proposed that allopolyploid cottons formed more than once, they suggested the best models of the potential D subgenome ancestral donor of allopolyploid cottons were D1 (G. thurberii), D3-d(G. davidsonii), D3-k (G. klotzschianum) and D5(G. raimondii), so they considered the allopolyploid cottons are polyphyletic (Kammacher, 1960; Sherwin, 1970; Johnson, 1975; Umbeck, 1985; Stewart, 1987; Da and Bertrand,1995). However, Most authors proposed that allopolyploid cottons formed only once, and that D5 was more similar to the D subgenome of allotetraploid cottons than other D genome diploids, they considered the allopolyploid cottons are monophyletic (Phillips,1963, 1964, 1966; Endrizzi et al., 1985; Wendel, 1989; Cronn et al.,1996; Seelanan et al., 1997; Small and Wendel, 1999, 2000; Wendel and Albert et al., 1992 ;Wendel et al., 1995; Wendel and Cronn, 2002; Admas, 2003).
We compared the GISH signals and DA values of (AD)1 and (AD)2 chromosomes generated by different diploid D genome species. The DA values of D6(G. gossypioides) were much higher than those of other D genome species both in (AD)1 and (AD)2, but the signals were distributed on both A and D subgenome chromosomes of (AD)1 and (AD)2, and the signal intensity was much weaker than that produced by other D genome species such as D3-d (G. davidsonii) and D4 (G. aridum) in (AD)1, or D1 (G. thurberi), D3-d (G. davidsonii) and G. raimondii (D5) in (AD)2. Therefore, the D6 genome species cannot be the D subgenome progenitor donor of (AD)1 and (AD)2.
The signal intensity in D3-d probe was more stronger than any other D genome species probes in GISH of (AD)1, and the DA value of D3-d probe was much higher than other D genome species too. This indicated that D3-d genome species has the strongest ability to recognize the D subgenome chromosomes of (AD)1, so we suggested that D3-d but not D5 was the possible D subgenome progenitor donor of (AD)1.
And on the other hand, the signal intensity in D5 probe was more stronger than any other D genome probes in GISH of (AD)2, and the DA value of D5 probe was also much higher than other D genome probes.This indicated that D5 genome species has the strongest ability to recognize the D subgenome chromosomes of (AD)2, so we thought that the D5 is the possible D subgenome progenitor donor of (AD)2.
Previous GISH studies had shown that diploid D1 and D3-d genome species was the D sub-genome progenitor donor of (AD)4 (G. mustelinum) and (AD)5 (G. darwinii) respectively (Wu et al., 2010; 2013), combined with the results of this experiment, allopolyploid cottons maybe formed with different D genome species as the progenitor donor, these results supported the hypothesis that allopolyploid cottons are polyphyletic.
3 Materials and Methods
We used allotetraploid cultivated cotton species (2n = 4x = 52) (AD)1 (G. hirsutum var. Zhongmiansuo 16) and (AD)2 (G. barbadense var. Xinhai 7) as target chromosomes. The Probes were made from diploid cotton (2n = 2x = 26) genomic DNA (gDNA), including all 3 A genome species G. herbaceum var. hongxing (A1), and G. arboreum var. shixiya 1 (A2), and G. herbaceum wild var. arfrium (A1-a); and all 13 D genome species, including G. thurberi (D1), G. armourianum (D2-1), G. harknessii (D2-2), G. davidsonii (D3-d), G. klotzschianum (D3-k), G. aridum (D4), G. raimondii (D5), G. gossypiodies (D6), G. lobatum (D7), G. trilobum (D8), G. laxum (D9), G. turneri (D10) and G. schwendimanii (D11). All of the plant materials are being taken from National Wild Cotton Germplasm Nursery in Sanya city, Hainan, China.
Total gDNA was extracted and purificated from immature leaves using the CTAB method (Song et al., 1999). The purified total gDNA was cut off with 120℃ 10 min, and examined with 0.8% agarose gel electrophoresis for their fragment size, which was generally appropriate in 300-600bp, and then tagged with markers. The gDNA probes were labeled with DIG-High-Prime labeling system (Roche Company, Germany), according to the standard operating procedures. The preparation of mitotic metaphase chromosomes which derived from the root tip cells and the procedure of GISH referenced to the method of Wang (Wang et al., 1999).
The hybridization signals were observed using a fluorescence microscope (Ziess Axioskop 2 plus). Images were captured by ISIS (in situ imaging system) software by adjusting their brightness and contrast. And also used this software to calculate the DA (distinguishing ability) value which was described by Markova et al (2007). And used Adobe Photoshop 7.0 software makes the plate.
Acknowledgment
This work was supported by the Exploring and Ultilization of Economic Crop Germplasm Resources(2013BAD01B03), the National Agriculture Research System of Cotton (CARS-18-36) and the Subproject of the Major Projects of Genetically Modified Organisms Breeding (2011ZX08005002-009) and also the Science and Technology Plan of the Jiangxi Province (20141BBF60011).
Adams K.L., Cronn, R., Percifield R., and Wendel J.F., 2003,Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing, Proc. Nat. Acad. Sci. USA, 100(8): 4649-4654
http://dx.doi.org/10.1073/pnas.0630618100 PMid:12665616 PMCid:PMC153610
Beasley J.O. , 1940, The origin of American tetraploid Gossypium species, Am. Nat., 74(7): 285-286
http://dx.doi.org/10.1086/280895
Cheng Z.K., Dong F.G., Langdon T., Ouyang S., Buell C.R., Gu M.H., Blattner F.R.,and Jiang J.M., 2002, Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon, Plant Cell,14(8):1691-1704
http://dx.doi.org/10.1105/tpc.003079 PMid:12172016 PMCid:PMC151459
Cronn R.C., Zhao X.P., Paterson A.H., and Wendel J.F., 1996, Polymorphism and concerted evolution in a tandemly repeated gene family: 5S ribosomal DNA in diploid and allopolyploid cottons, J. Mol. Evol., 42(6): 685-705
http://dx.doi.org/10.1007/BF02338802 PMid:8662014
Cui X.L., Liu F.,Liu Y.L.,Zhou Z.L.,Zhao Y.Y.,Wang C.Y.,Wang X.X.,Cai X.Y.,Wang Y.H.,Meng F.,Peng R.H.,and Wang K.B., 2015, Construction of cytogenetic map of Gossypium herbaceum chromosome 1 and its integration with genetic maps, Mol. Cytogenetic, 8:2-10
http://dx.doi.org/10.1186/s13039-015-0106-y PMid:25628758 PMCid:PMC4307992
Da Rocha P.S.C.F., and Bertrand H., 1995, Structure and comparative analysis of the rDNA intergenic spacer of Brassica rapa:Implication for the function and evolution of the Cruciferae spacer, Eur. J. Biochem, 229(2): 550-557
http://dx.doi.org/10.1111/j.1432-1033.1995.tb20497.x PMid:7744079
Dejooded D.R., and Wendel J.F., 1992, Genetic diversity and origin of the Hawaiian Islands cotton,Gossypium tomentosum, Amer. J. Bot., 79: 1311-1319
http://dx.doi.org/10.2307/2445059
Endrizzi J.E., Turcotte E.L., and Kohel R.J., 1985, Genetics, cytogenetics, and evolution of Gossypium, Adv. Genet.,23:271-375
http://dx.doi.org/10.1016/S0065-2660(08)60515-5
Fryxell P.A., 1992, A revised taxonomic interpretation of Gossypium L.(Malvaceae), Rheedea,2(2): 108-165
Gan Y., Liu F., Chen D.,Wu Q., Qin Q.,Wang C., Li S., Zhang X.,Wang Y.,and Wang K., 2013, Chromosomal Locations of 5S and 45S rDNA in Gossypium Genus and Its Phylogenetic Implications Revealed by FISH, PloS one, 8(6): e68207
http://dx.doi.org/10.1371/journal.pone.0068207 PMid:23826377 PMCid:PMC3695094
Gerstel D.U., 1953, Chromosomal translocations in interspecific hybrids of the genus Gossypium,Evolution, 7: 234-244
http://dx.doi.org/10.2307/2405734
Jiang J.M., and Gill B.S.,2006, Current status and the future of fluorescence in situ hybridization (FISH) in plant genome research, Genome, 49(9):1057-1068
http://dx.doi.org/10.1139/g06-076 PMid:17110986
Johnson B.L., 1975, Gossypium palmeri and a polyphyletic origin of the New World cottons, Bull Torrey Bot. Club, 102(6): 340-349
http://dx.doi.org/10.2307/2484760
Kammacher P., 1960, Observations cytologiques sur deux hybrides F1 entre especes cultivees tetraploides de cotonniers et l’espece diploide sauvage Gossypium raimondii, UlB. Rev. Cytol. Biol. Veg., 22(5): 1-31
Liu B., and Wendel J.F., 2001, Intersimple sequences repeat (ISSR) polymorphisms as a genetic marker system in cotton,Molecular Ecology Notes, 1(3):205-208
http://dx.doi.org/10.1046/j.1471-8278.2001.00073.x
Liu S.H., Wang K.B., Song G.L., Wang C.Y., Liu F., Li S.H., Zhang X.D., and Wang Y.H., 2005, Primary investigation on GISH-NOR in cotton, Chinese Science Bulletin, 50(5): 425-429
http://dx.doi.org/10.1007/BF02897457 http://dx.doi.org/10.1360/982004-582
Markova M., Michu E., Vyskot B., Janousek B., and Zluvova J., 2007, An interspecific hybrid as a tool to study phylogenetic relationships in plants using the GISH technique, Chromosome Research, 15(8):1051-1059
http://dx.doi.org/10.1007/s10577-007-1180-8 PMid:18075777
Melo C.A., Silva G.S., and Souza M.M., 2015, Establishment of the genomic in situ hybridization (GISH) technique for analysis in interspecific hybrids of Passiflora,Genet. Mol. Res., 14(3):2176-2188
http://dx.doi.org/10.4238/2015.March.27.4 PMid:25867365
Mukai Y.,Nakahara Y.,and Yamamoto M., 1993, Simultaneous discrimination of the three genomes in hexaploid wheat by multicolor fluorescence in situ hybridization using total genomic and highly repeated DNA probes, Genome,36(3):489-494
http://dx.doi.org/10.1139/g93-067 PMid:18470003
Nemeth C., Yang C.Y., Kasprzak P., Hubbart S., Scholefield D., Mehra S., Skipper E., and King J., 2015, Generation of amphidiploids from hybrids of wheat and related species from the genera Aegilops, Secale, Thinopyrum, and Triticum as a source of genetic variation for wheat improvement, Genome ,58 (2): 71-79
http://dx.doi.org/10.1139/gen-2015-0002 PMid:26053312
Phillips L.L., 1963, The cytogenetics of Gossypium and the origin of new world cotton, Evolution,17:460-496
http://dx.doi.org/10.2307/2407096
Phillips L.L., 1966, The cytology and phylogenetics of the diploid species of Gossypium, Amer. J. Bot., 53(4): 328-335
http://dx.doi.org/10.2307/2439872
Phillips L.L., 1964, Segregation in new alloploids of Gossypiumâ…¤. Mutivalent formation in new world×Asiatic and new world×wild American hexaploids,Ame. J. Bot., 51(3):324-329
Rayburn A.L., and Gill B.S.,1985, Use of biotin labeled probes to map specific DNA sequences on wheat chromosomes, J. Hered., 76:78-81
Seelanan T., Schnabel A., and Wendel J.F., 1997, Congruence and consensus in the cotton tribe, Syst. Bot., 22(2): 259-290
Sherwin K.H., 1970, Winds across the Atlantic-possible African origins for some Pre-Columbia New World CULTIGENS, Res. Rec. Univ. Mus. S. 111. Univ. Mso-Am. Stand, 6:1-33
Small R.L., and Wendel J.F., 1999, The mitochondrial genome of allotetraploid cotton (Gossypium L.), J. Hered., 90(1): 251-253
http://dx.doi.org/10.1093/jhered/90.1.251 PMid:9987935
Small R.L., Ryburn J.A., and Cronn R.C., Seelanan T., Wendel J. F., 1998, The tortoise and the hare: Choosing between noncoding plastome and nuclear Adh sequences for phylogeny reconstruction in a recently diverged plant group. Am. J. Bot., 85(9): 1301-1315
http://dx.doi.org/10.2307/2446640 PMid:21685016
Small R.L., and Wendel J. F., 2000, Copy number lability and evolutionary dynamics of the Adh gene family in diploid and tetraploid cotton (Gossypium), Genetics, 155(4):1913-1926
Snowdon R.J., Kohler W., Friedt W., and Kohler A.,1997, Genomic in situ hybridization in Brassica amphidiploids and interspecific hybrids, Theor. Appl. Genet., 95(8): 1320-1324
http://dx.doi.org/10.1007/s001220050699
Song G.L., Cui R.X., Wang K.B.,Li S.H.,Zhang J.F., and Guo J.H., 1999, Analysis of genetic diversity of Australian species of Gossypium using RAPD, Cotton Science, 11(2): 65-69
Stewart J.M., Craven L.A., and Fryxell P.A., 1987, Gossypium germplasm from Australia, Plant Genetic Resources Newsletter, 69: 44-47
Tang S., Li X., and Jia X., 2000, Genomic in situ hybridization (GISH) analyses of thinopyrum intermedium, its partial amphiploid zhong 5, and disease-resistant derivatives in wheat. Theor. Appl. Genet., 100(3): 344-352
http://dx.doi.org/10.1007/s001220050045
Umbeck P.F., and Stewart J.M., 1985, Substitution of cotton cytoplasms from wild diploid species for cotton germplasm improvement, Crop Sci., 25(6): 1015-1019
http://dx.doi.org/10.2135/cropsci1985.0011183X002500060028x
Wang K., Guan B., Guo W.Z.,Zhou B.L., Hu Y.,Zhu Y.C., and Zhang T.Z.,2008, Completely distinguishing individual A-genome chromosomes and their karyotyping analysis by multiple bacterial artificial chromosome–fluorescence in situ hybridization, Genetics, 178(2): 1117-1122
http://dx.doi.org/10.1534/genetics.107.083576 PMid:18287408 PMCid:PMC2248359
Wang K., Yang Z.J., Shu C.S., Hu J.,Lin Q.Y.,Zhang W.P.,Guo W.Z.,and Zhang T.Z., 2009, Higher axial-resolution and sensitivity pachytene fluorescence in situ hybridization protocol in tetraploid cotton, Chromosome Research, 17(8): 1041-1050
http://dx.doi.org/10.1007/s10577-009-9085-3 PMid:19844799
Wang C.Y., Wang K.B., Wang W.K., Li M.X., Song G.L., Cui R.X., Li S.H., Zhang, X.D., and Zhang J.M., 1999, Protocol of cotton FISH of somatic chromosomes with gDNA as probes, Cotton Science, 11(2):79-83
Wang K.B., Du X.M., and Song G.L.,2004, The present situation and development of the cotton germplasm creation, Plant genetic resources, 5(supplement): 23-28
Wang K.B., Song G.L., Wang C.Y., Liu F., Li S.H., Zhang X.D., and Wang Y.H., 2008, FISH-based karyotype of Gossypium herbaceum generated with 45s rDNA and gDNA of Gossypium raimondii as probes, Cotton Science, 20(4): 264-273
Wang K.B., Wang W.K., Wang C.Y., Song G.L., Cui R.X., Li S.H., and Zhang X.D., 2001, Studies of FISH and karyotype of Gossypium barbadense, Acta Genetica Sinica, 28(1): 69-75
Wu Q., Cheng H., Liu F., Wang S.F., Wang C.Y., Song G.L., Li S.H., Zhang X.D., Wang Y.H., Ma Z.Y., and Wang, K.B., 2010, Screen of FISH marker of chromosomes at Gossypium D genome species, Chinese Science Bulletin, 55(21): 2099-2105
Wu Q., Liu F., Li S., Song G., Wang C., Zhang X., Wang Y., Stelly D., and Wang K., 2013,Uniqueness of the Gossypium mustelinum genome revealed by GISH and 45S rDNA FISH, J. Integr. Plant. Biol., 55(7):654-662
http://dx.doi.org/10.1111/jipb.12084 PMid:23758934 PMCid:PMC3810724
. PDF(717KB)
. FPDF
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Xiao Shuiping
. Wang Kunbo
. Yang Lei
. Ke Xingsheng
. Wang Chunying
. Liu Xinwen
. Sun Liangqing
. Yang Shaoqun
. Liu Fang
. Chen Yi
Related articles
. Gossypium
. G.hirsutum L.
. G.barbadense L.
. Progenitor
. GISH
. GISH-NOR
Tools
. Email to a friend
. Post a comment
.png)

.png)


