Assessment of Genetic Diversity and Population Structure in a Selected Germplasm Collection of 292 Jute Genotypes by Microsatellite (SSR) Markers 
2. Global-Theme Biotechnology, International Crops Research Institute for Semi-Arid Tropics (ICRISAT), Patancheru, Hyderabad, 502324, India
3. Biotechnology Unit, Division of Crop Improvement, Central Research Institute for Jute and Allied Fibres (ICAR), Barrackpore, 743101 West Bengal, India
 Author
Author  Correspondence author
Correspondence author
Molecular Plant Breeding, 2012, Vol. 3, No. 2 doi: 10.5376/mpb.2012.03.0002
Received: 26 Dec., 2011 Accepted: 02 Feb., 2012 Published: 15 Mar., 2012
Sumana Banerjee et al., 2012, Assessment of Genetic Diversity and Population Structure in a Selected Germplasm Collection of 292 Jute Genotypes by Microsatellite (SSR) Markers, Molecular Plant Breeding, Vol.3, No.2 11-25 (doi: 10.5376/mpb.2012.03.0002)
Genetic diversity within the available jute germplasm has not been characterised in detail. Therefore, using 172 SSRs developed in our laboratory, we assessed genetic diversity and population structure in 292 genotypes (including both indigenous and exotic accessions) of two cultivated species (C. capsularis L. and C. olitorius L.). Altogether, as many as 596 alleles (3.46 alleles per locus) were detected. The average values of PIC calculated over all the loci did not differ significantly in the two species (0.198 in C. capsularis and 0.203 in C. olitorius). In C. capsularis, 112 markers (2.55 alleles per locus) and in C. olitorius 140 markers (2.75 alleles per locus) were polymorphic. During both distance based cluster analyses and model based structure analyses, most of the indigenous and exotic genotypes of the two species clearly delineated into separate groups. Basic statistics (Ne, I and He) revealed that the exotic genotypes were slightly more diverse than the indigenous genotypes in C. olitorius while the reverse was true in case of C. capsularis. In each of the two species, low level of population differentiation (Fst) was observed between the indigenous and exotic genotypes, which are also congruent with the results of AMOVA and Nei’s genetic distance, suggesting incidence of gene flow through germplasm exchange across countries. Seven pairs of most divergent genotypes of the two species were identified by pair-wise genetic distance analysis which could be useful for development of jute genotypes with improved fibre yield and fibre quality traits.
The genus Corchorus (known as jute) belongs to the family Sparrmanniaceae, which has recently been carved out of the family Malvaceae sensu lato (Heywood et al., 2007). It comprises 50~60 species (Mahapatra and Saha, 2008), which are distributed in the tropical lowland areas (Edmonds 1990; Benor et al., 2010), although only two diploid (2n = 2x = 14) species, namely C. capsularis L. (the white jute) and C. olitorius L. (the tossa jute/Jew’s mellow) are cultivated for fibre yield. These two species differ among themselves in terms of their centres of origin (Basu et al., 2004). C. capsularis is believed to have originated in the Indo-Burma region, including south China, while C. olitorius is believed to have originated in Africa (Kundu, 1951). The two species also differ with respect to their genome sizes. Recent studies have suggested that the genome size of C. olitorius is greater (~324 Mb to ~449 Mb) than the genome size of C. capsularis (~280 Mb to 392 Mb) (Benor et al., 2011; Sarkar et al., 2011).
Jute is a natural fibre-crop, which is second only to cotton in its importance. Its cultivation is restricted to the Indian subcontinent and parts of south-east Asia. India is also the largest producer of jute contributing 58% of jute fibre to the global jute production followed by Bangladesh with 33% production. Jute is mainly cultivated in eastern parts of India and north Andhra Pradesh covering 9.37 lakh hectares area. The current average jute production in India is 109.79 lakh bales per annum (1 bale = 180 kg) (http://www.cag.gov.in/html/reports/commercial/2010-11_10PA/chap10.pdf). In recent years the importance of jute crop has increased further due to its use in diversified value-added industrial products and packaging materials (Mondal, 2000).
The yield and quality of jute fibre need to be improved further through redesigning of breeding programs to allow its successful competition with widely and abundantly used synthetic fibres. However, only very limited number of indigenous and exotic jute accessions representing only a narrow range of genetic variability have been exploited so far in jute breeding programs (Roy et al., 2006). Early success in varietal development of tossa jute was achieved through utilization of an African genotype “Sudan green”, while induced mutations were the major source for varietal development of white jute. Until 2010, only 16 varieties of C. capsularis and 15 varieties of C. olitorius were released in India, mainly following intra-specific hybridization (Kar et al., 2010). Therefore, there is a need for the use of newer and diverse germplasm for jute improvement programs.
C. capsularis has fine fibre with slightly weak strength. It is relatively resistant to flood and drought and also susceptible to diseases and pests. In contrast, C. olitorius has stronger and lustrous fibre and is also more resistant to diseases and pests, although it is susceptible to abiotic stresses like flood and drought. Therefore, ideally a desirable approach for jute improvement would be to combine the contrasting features of the two species into a single genotype. However, these two species are cross incompatible precluding inter-specific hybridization for combining the desirable traits (Patel and Datta, 1960; Swaminathan et al., 1961). Therefore, only intra-specific hybridization involving diverse germplasm may be used for producing improved varieties belonging to each of these two commercially important species of jute. Although a large collection of 939 accessions of C. capsularis and 1 647 accessions of C. olitorius is available at Central Research Institute for Jute and Allied Fibres (CRIJAF), Barrackpore, Kolkata, India, but systematic and planned efforts have seldom been made to study genetic diversity in the available germplasm of the two cultivated species. Also, genetic diversity in jute was studied in the past using only morpho-physiological traits such as plant height, harvest index, cambial activity and fibre strength (Palit et al., 1996). These traits are limited in number and are often influenced by the environment, and therefore, may not be suitable for correct assessment of the genetic diversity. This limitation can be largely overcome by the use of molecular markers, which are unlimited in number and are not influenced by the environment.
The molecular markers have been widely used for the study of genetic diversity in almost all crops (Prasad et al., 2000; Malysheva-Otto et al., 2006; Mazzucato et al., 2008; Wen et al., 2009; Ming et al., 2010; Zarkti et al., 2010; Mir et al., 2011). In jute, molecular markers like RAPDs, chloroplast-SSRs, gSSRs, ISSRs, and AFLPs have been used to assess genetic diversity in both cultivated and wild species (Qi et al., 2003a; 2003b; Hossain et al., 2002a; 2003b; Basu et al., 2004; Roy et al., 2006; Haque et al., 2007; Mir et al., 2008a; Mir et al., 2008b; Akter et al., 2008; Huq et al., 2009). However, these studies were carried out on rather small collections of up to 81 genotypes of cultivated and wild species giving only very limited idea about the genetic diversity available in the gene pool of the two cultivated species of jute. The present study on genetic diversity and population structure was conducted using a relatively larger and more diverse set of 292 jute genotypes (152 genotypes of C. capsularis and 140 genotypes of C. olitorius) from both indigenous and exotic collections with the help of 172 genomic SSRs.
1 Results
1.1 SSR polymorphism
Genotyping of all the 292 jute genotypes including 152 genotypes of C. capsularis and 140 genotypes of C. olitorius was carried out using a set of 167 SSR primers which identified 172 polymorphic SSR loci carrying 596 alleles (3.45 alleles per locus, range 2~7 alleles). Out of the 172 polymorphic loci, 8 loci (4.65%) exhibited only inter-specific polymorphism and were bi-allelic, one allele for each of the two species and 93 SSR loci (~54%) accounted for 150 alleles shared by the both species. The remaining 71 loci (41.27%) exhibited both inter and intra-specific polymorphism between the two species (Supplementary Table 1). Out of a total 596 alleles, 179 alleles were unique to C. capsularis and 267 alleles were unique to C. olitorius. The PIC per SSR locus calculated over all SSR loci did not differ much in the two species (0.198 vs 0.203; Table 1).
|
Table 1 SSR polymorphism estimated over all SSR markers among the two species of Corchorus |
All the 172 polymorphic SSR loci used in the present study were amplified in C. olitorius, but cross-species amplification of only 156 SSRs (90.69%) showing expected size of fragments was observed within C. capsularis (Supplementary Table 1). In C. capsularis, a set of 112 loci were polymorphic carrying 286 alleles (2.55 alleles per locus) and in C. olitorius, 140 loci were polymorphic carrying 385 alleles (2.75 alleles per locus) (Table 1; Table 2; Figure 1). The list of polymorphic loci with range and mean of PIC values are presented in Table 2. A fairly large fraction of the polymorphic SSR loci were unique (24 loci in C. capsularis and 52 loci in C. olitorius) to each of the two species of jute (Table 2).
.png) Table 2 Detailed genetic information of polymorphic SSRs in the two cultivated jute species |
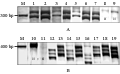 Figure 1 A representative figure showing polymorphism due to (A) SSR MJM212 in C. capsularis and (B) SSR MJM609 in C. olitorius |
1.2 Rare alleles
Out of 172 SSR loci, 118 loci had 157 rare alleles, each allele having a frequency of <5% (Supplementary Table 1). Out of 157 rare alleles, 25 were available in both the species; 59 rare alleles were unique to C. capsularis and 73 rare alleles were unique to C. olitorius (Table 1). Some of the above rare alleles were genotype specific (i. e. alleles were detected in only one genotype; Table 3). In C. capsularis, two exotic genotypes (one each from Nepal and Sri Lanka) and only one indigenous genotype showed genotype-specific alleles; similarly, in C. olitorius, three genotypes of exotic origin and four genotypes of indigenous origin exhibited genotype-specific alleles (for details see Table 3).
 Table 3 Details of genotype specific alleles identified among the genotypes of C. capsularis and C. olitorius |
1.3 Genetic diversity studied by cluster analysis
The un-weighted neighbour-joining (UNJ) dendrograms for both the species together and separately for each species are presented in Figure 2. In the combined dendrogram, the genotypes of the two species were largely grouped in two separate clusters, cluster â… and cluster â…¡ (Figure 2a). However, 14 (out of 152) genotypes of C. capsularis were grouped with genotypes of C. olitorius and similarly 19 (out of 140) genotypes of C. olitorius were grouped with genotypes of C. capsularis (circled regions in Figure 2a). The dendrogram for C. capsularis (Figure 2b) had three clusters with cluster â…¡ having two sub-clusters i.e. â…¡a and â…¡b. Cluster â… , cluster â…¡a and cluster â…¢ contained only indigenous jute genotypes except the two exotic genotypes (CIJ 091 from Thailand and CIJ 130 from China) in cluster â…¢. In cluster â…¡b, all jute genotypes were exotic except for the four indigenous genotypes. The dendrogram for C. olitorius also has three clusters, namely â… , â…¡ and â…¢ (Figure 2c). The cluster â… included only exotic genotypes and the cluster â…¢ contained indigenous genotypes except for a solitary genotype OIJ 277 from Nepal. However, cluster â…¡ included both indigenous and exotic genotypes.
 Figure 2 Un-weighted neighbor-joining dendrograms of jute genotypes |
The principal coordinate analysis (PCA) was also performed to study the grouping of genotypes which is primarily explained by the first two principal coordinates. The PCA plots are presented in Figure 3. In the combined analysis, the two species formed two separate and clear groups in PCA plot (Figure 3a), where the first two coordinates explained 67.25% and 9.89% of the total variation. In the independent PCA analyses for each of the two species also, the first two coordinates explained 29.76% and 19.74% of the total variation in C. capsularis and 33.08% and 21.48% of the total variation in C. olitorius. The grouping of genotypes in all the three PCA plots was largely in agreement with those observed in UNJ dendrograms. In the PCA plots of the two species, three groups each corresponded to the three clusters of the UNJ dendrograms (Figure 3b and 3c).
 Figure 3 Principal coordinate analysis based on SSR markers showing distribution of jute genotypes |
1.4 Analysis of molecular variance (AMOVA)
The results of AMOVA indicated that proportion of variation between the two jute species accounted for most (63%) of the molecular variance and only 37% of the variance accounted for variation within each species. The genotypes of both C. capsularis and C. olitorius were sub-grouped according to their country of origin into indigenous and exotic collections and for partitioning the genetic variation among the collections of genotypes of both the species, AMOVA was performed (Table 4). The variation among the genotypes within indigenous and exotic collections in each of the two species was 7-9 times higher than the variation between the indigenous and exotic collections (Table 4).
|
Table 4 AMOVA for all jute genotypes and separately for the genotypes in two species based different set of SSR primers |
1.5 Genetic differentiation within indigenous and exotic collections of genotypes in the two jute species
A comparison of diversity statistics within the indigenous and exotic collections of genotypes separately for the two species is presented in Table 5. The values of I and He (except Ne) were higher in the indigenous genotypes than in the exotic genotypes belonging to C. capsularis. In C. olitorius the exotic genotypes showed slightly higher values of Ne, I and He than the indigenous genotypes (Table 5). The above study revealed that the exotic genotypes were more diverse than the indigenous genotypes of C. olitorius and the reverse situation was observed in the case of C. capsularis.
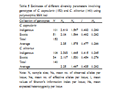 Table 5 Estimates of different diversity parameters involving genotypes of C. capsularis (152) and C. olitorius (140) using polymorphic SSR loci |
Further analyses were conducted on specific alleles which were present in either indigenous or exotic collection; these are described as private alleles (Table 6; Table 7). Moreover, in C. capsularis 24.13% of private alleles and in C. olitorius 25.58% of private alleles (Table 7) showed allele frequency greater than 6%. The number of private alleles for exotic collection was higher in C. olitorius than in C. capsularis. Also, only few SSR loci were shared between indigenous and exotic collections which are higher in C. olitorius (Table 6).
|
Table 6 Alleles specifically present in indigenous and exotic collection of genotypes of C. capsularis and C. olitorius |
|
Table 7 List of SSR loci showing private alleles with allele frequency >6% in indigenous and exotic collections of the two species of jute |
The pair-wise values for unbiased genetic distance (GD), genetic identity (GI), population differentiation (Fst) and gene flow (Nm) between the indigenous and exotic collections of both the species are presented in Table 8. The data clearly showed that there is low genetic distance (GD) and population differentiation (Fst) between the indigenous and exotic genotypes of both the species. However, population differentiation between the indigenous and exotic genotypes was slightly higher in C. capsularis, than in C. olitorius. Correspondingly, higher level of gene flow existed between the indigenous and exotic collection in C. olitorius (Nm=5.206) than in
. PDF(1515KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Sumana Banerjee
. Moumita Das
. Reyazul Rouf Mir
. Avijit Kundu
. Niladri Topdar
. Debabrata Sarkar
. Mohit K Sinha
. Harindra S Balyan
. Pushpendra K Gupta
Related articles
. Corchorus capsularis
. Corchorus olitorius
. Genetic diversity
. Population structure
Tools
. Email to a friend
. Post a comment


