2. College of Agriculture & Biotechnology, China Agricultural University, Beijing, 100193, P.R. China
3. College of Forestry, Northeast Forestry University, Harbin, 150040, P.R. China
* These authors contributed equally to this study
 Author
Author  Correspondence author
Correspondence author
Molecular Plant Breeding, 2012, Vol. 3, No. 3 doi: 10.5376/mpb.2012.03.0003
Received: 22 Feb., 2012 Accepted: 28 Mar., 2012 Published: 01 Apr., 2012
Ji et al., 2012, Genetic Analysis of ESTs Generated from Pericarp of Wine Grape (V. amurensis) at Veraison, Molecular Plant Breeding, Vol.3, No.3 26-36 (doi: 10.5376/mpb.2012.03.0003)
In this study, we constructed the cDNA library of grape berry pericarp at veraison from wine grape ‘Shuang Feng’ (V. amurensis), and generated total 974 ESTs, representing the independent cDNA in length of around 392,672 bp, of which 710 unigenes were clustered into 97 contigs and 613 singletons based on quality of EST sequence and optimum assembly. 64.65% clones exhibited high homology to previously deposited nucleotides and/or polypeptide sequences, while the proportion of ESTs with no any significant homology in database reached 25.63%. Analysis of the unigenes resulted in the assignment of 9 specific functional classes in this research. Putative functions were identified for about 74.13% of the unigenes. By assigning them to specific function classes, we were able to highlight characteristics among different ripening phases. GO classification was performed to define the relationship among genes expressing in pericarp of wine grape berry, which were involved in three aspects including cell component, biological process and molecular function. Expression analysis of these ESTs were used to characterize potential roles of novel genes with respect to berry ripening and composition of wine grape.
Grapevine is the most valuable horticultural crop worldwide. Grape berries are processed into wine, produced commercially for fresh consumption, dried into raisins, processed into non-alcoholic juice and distilled into spirits (Lund et al., 2008). As one of the most important wild germplasm resources, V. amurensis (Amur grape) is widely distributed in China, Korean and Japan (Ha et al., 2009). It is very resistant to frost, withstanding temperature as low as -40℃, but it is not tolerant to drought. The roots, vine and leaves of the Amur grape are used as therapeutic agents for disease treatment according to the theory of traditional Chinese medical science. By hybridization with the V. vinifera native in Europe, several valuable cultivars resistant to low temperature have been produced (Weidner et al., 2007). Figure 1 Agarose gel electrophoresis of total RNA
Veraison is the period before berry ripening, during which dramatic changes of berry occur. The grapevine is considered to be a non-climacteric fruit, and follows the trace of growth of a double-sigmoid curve. The two successive phases both last around 6 weeks with similar amplitudes (Terrier et al., 2001). After fruit set, cell divisions and expansion result in the rapid growth of the berry. The following stage is a lag phase with little or no growth of berry. Subsequently, the second growth phase called veraison occurs, in which a series of physiological and biochemical changes take place. The berry becomes soft with a rapid increase in the level of hexoses in the berry vacuole and increase in berry volume. The organic acid content decreases while the level of soluble sugars increases. The chlorophyll breaks down and the color starts to turn. Certain secondary compounds involved with fruit flavor and aroma are formed (Coombe, 1992; Fillion et al., 1999; Davies and Robinson, 2000; Terrier et al., 2001).
Expressed sequence tags (ESTs) are one of the most useful and efficient tools in function genomics and comparative genomic studies. The worldwide grape EST projects were initiated several years ago and made a significant progress so far. In 2001, there were less than 400 V. vinifera ESTs in GenBank (da Silva et al., 2005; Moser et al., 2005), with large margins of increase from then on, this number has reached 352 730 ESTs according to the Release 7.0 (April 17, 2010; http://compbio.dfci.harvard.edu/cgi-bin/tgi/gimain.pl?gudb=grape) of the TIGR Grape Gene Index, which consists of 34 154 putative different transcripts and 31 813 singleton ESTs. Ablett et al (2000) fulfilled the V. vinifera sequences analysis of 2 479 ESTs and 2 438 ESTs from Chardonnay berry tissue and leaf respectively, revealing that 2 330 distinct sequences were matched with the non-redundant protein database. While Terrie et al (2001) analyzed the V. vinifera L. berries (cv. Shiraz) at various development stages, and 275 ESTs sequenced from their 3' ends were generated. Pacey-Miller et al (2003) constructed a grape bud cDNA library with 4 270 ESTs sequenced, indicating that gene expression in the buds is high even in the dormancy process. More recently, Moser et al (2005) carried out the sequence project with 8 147 ESTs generated from six different grape organs and 405 SSRs were identified. The available resource of EST sequences and annotations were organized in the IMAP-database. Peng et al (2007) constructed 11 cDNA libraries usd variety of tissues from two grapevine (V. vinifera L.) cultivars, Cabernet Sauvignon (wine grape) and Muscat Hamburg (table grape). 77 583 high quality ESTs were generated and analyzed, with 2 725 novel grape unigenes discovered, which may play the crucial roles in grape berry development and regulation of berry composition important for wine and table grape quality.
As mentioned above, in contrast with V. vinifera L., the molecular biological research of V. amurensis is limited. The disease-resistance genes and the related genes of anthocyanin biosynthesis pathway have been cloned, such as F3'H, 3GT, and so on (Liu et al., 2009), but the largest-scale EST sequencing project has a great potential for further research aiming to discover new genes and regulation mechanism of key enzymes. As the raw material for wine making, selecting the right ripening stage of the V. amurensis berry is critical for its use in the wine-industry. Therefore, we should focus on the berry development, so as to elucidate the occurrence and process of the changes, as well as the factors influencing it. In this research we constructed the cDNA library of V. amurensis pericarps at veraison, illuminate the gene functions through analyzing the gene expression and finding out novel genes after veraison.
1 Results and Analysis
1.1 RNA extraction
The modified CTAB protocol was efficient for high-quality RNA extraction from V. amurensis pericarp at veraison stage. The concentration of total RNA was 1.256 μg/μL, with the A260:A280 and A260:A230 ratios being 1.90 and 2.10 respectively. RNA examined by electrophoresis on 1.1% agarose gel showed the intact 28S and 18S rRNA bands with little smearing, indicating that little or no RNA degradation occurred during extraction (Figure 1). These parameters demonstrated a high quality of the total RNA that satisfied the needs of the cDNA library construction.
![]()
1.2 cDNA library quality evaluation
With the inverse transcription, the first-strand cDNA, which was used as component in the double-strand cDNA synthesis by LD-PCR was synthesized. Examined by electrophoresis on 1.1% agarose gel (Figure 2), the ds-cDNA size distribution appeared to have the range of 200~2 000 bp, with moderately strong smear of cDNA ranging from 500~1 000 bp, indicating that some genes are highly expressed in this period.
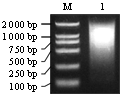 Figure 2 Agarose gel electrophoresis of ds cDNA from V. amurensis |
After the drip procedure by the CHROMA SPIN-400 Column, the 7~10 fractions containing cDNA with lengther no longer than 500 bp was collected (Figure 3). To obtain a library of the desired complexity, three separate ligations were performed for an optimal ratio of cDNA to vector. The results showed that the ligation with the ratio of 1:1.5 could produce high numbers of clones that would be pooled to form the original, unamplified library.
Figure 3 Agarose gel electrophoresis of V. amurensis ds cDNA after fractionation |
The library qualification evaluation showed that the titer of primary cDNA library was 1.12×106 pfu/mL and the titer of amplified library was 2.82×109 pfu/mL. Furthermore, the percentage of recombination was about 100%, and the fragment size of insert was 0.5~2.0 kb, with the average insert size of 0.80 kb (Figure 4). The results made clear that the cDNA library of V. amurensis veraison pericarp constructed by SMARTTM was successful and had high quality.
Figure 4 PCR detection of inserts size from V. amurensis cDNA library on agarose gel |
1.3 cDNA library characterization
The size-distribution of 1 009 individual clones derived from the library constructed at veraison stage was analyzed. The average total length was 550.14 bp without any treatment. After vector sequence suppression, the EST sequence was 407.17 bp long on average, in which there were 11 vector sequences. The Poly A tails were cut off, and then the ESTs with lengths less than 100 bp, 35 sequences in total were removed. Finally, 974 high quality ESTs were obtained, representing as much as 96.53% of the total sequences. The percentage of vector sequences and low quality sequences was 3.74%. The total valid length ranged between 100 bp to 595 bp, and the mean valid length was 403.15 bp. The EST sequences described here have been deposited into GenBank dbEST database under accession numbers GW666520~GW667493. 710 unigenes were obtained using the phrap software, including 97 contigs and 613 singletons. The valid lengths of unigene sequences ranged from 102 bp to 1 024 bp with the average length of 431.35 bp. Redundancy of individual clones in the veraison library was 27.10%, which turned out to increase during development (Table 1). Davies and Robinson (2000) identified a few genes known as Grape Ripening Induced Proteins (GRIP), the over-expression of which led to the increased redundancy.
 Table 1 General characteristics of Vitis amurensis veraison pericarp ESTs |
1.4 Expression abundance of EST sequences
The EST numbers of the same gene obtained by randomly sequencing represent the expression abundance of the gene in the specific tissues and organs to a certain extent. Generally, the cell specific genes are expressed at a high level, while the expression of most house-keeping genes is in low abundance. There were 16 repeats in high quality ESTs, which represented 1.64%. When analyzed using BLAST after removing the repeats, there were 17 genes with high abundance (expression frequency ≥5), 80 medium abundance genes (expression frequency 2~5), and the rest were all in low abundance (Figure 5). The results indicated that the majority of genes were expressed in low abundance.
 Figure 5 The frequency of occurrence of uniseqs derived from spliced EST |
1.5 Grape pericarp ESTs with similarities to known sequences
Comparing with non-redundant database on NCBI website using BLASTX, the ESTs were divided into different groups based on their functional annotations. The homologies of ESTs have significant similarity in gene structure and functional characterization. There were 710 unigenes annotated. Among the 710 unigenes analyzed, 64.65% showed strong similarity (BLAST score >80) with known or unknown functions in the public database, and 9.44% showed marginal similarity (80> BLAST score >40). Therefore, 528 had either strong or weak homology to previously identified genes, which represented 722 ESTs with the percentage of 74.13% in the valid EST sequence. Of the 528 unigenes, 270 were characterized with known functions or putative functions, and there were 258 annotated with unknown functions. 25.87% of the analyzed unigenes did not match with any sequences in database. Some of these genes could actually be the key enzymes in biosynthesis pathway or the critical factors for controlling veraison, which is still deeply investigated and not fully understood at the molecular level (Table 2).
 Table 2 Selected set of specific sequences and their putative functions |
1.6 Species resource of homologous sequences
The species resource of homologous sequences were matched in a databasederived from 24 species. The V. vinifera had the best matching result with 69.1% homologous genes. Populus trichocarpa showed the second best result followed by Ricinus communis, V. hybrid cultivar, Nicotiana tabacum, V. pseudoreticulata, Sorghum bicolor. The other 17 species derived from Solanum tuberosum, Coffea Arabica, Arabidopsis thaliana, Olea europaea, Cucumis sativus, Zea mays and so on made up 2.39% of the total unigenes (Table 3).
 Table 3 Statistics of organism origin of matched function homologues |
1.7 Go classification
For gene ontology (GO) annotation, each sequence was afforded at least one GO term. The gene with different molecular function, biological process or cell component was counted repeatedly and respectively. 199 genes were classified as cell component (Figure 6), while313 genes are classified as molecular function, and 341 as biological process (Figure 7; Figure 8). Additionally, the number of ESTs can reflect the copy quantities of gene expression, which relate the proportion to the frequency of gene expression indirectly. As shown in the three pie charts (Figure 6; Figure 7; Figure 8), there is a high percentage of ESTs related to cell, antioxidant activity and cellular process in three different fields, respectively.
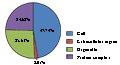 Figure 6 Gene ontology cell component classification of annotated genes |
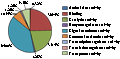 Figure 7 Gene ontology molecular function classification of annotated genes |
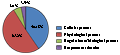 Figure 8 Gene ontology biological process classification of annotated genes |
For the purpose of better understanding the overall mechanisms in the pericarps under investigation, Gene Ontology (GO) annotations were performed and their correlated MIPS functional categories assigned 710 unigenes to 9 classes of biological function (Figure 9). In accordance with the biological roles of products encoded by genes, the nine classes included energy/metabolism (14.79%), protein synthesis (7.46%), signal transduction (2.82%), cell disease/defense (2.54%), transport (3.67%), photosynthesis (0.70%), transcription (2.54%), protein activity regulation (1.69%) and cell structure (1.83%). The most represented functional classes appeared to be energy/metabolism, which was followed by the protein synthesis and transport; however, the percentage of independent clones matching with sequences of putative or hypothetical protein without any predicted function was 36.34%. Therefore, dramatic changes at veraison are mainly attributed to energy/metabolism and protein synthesis.
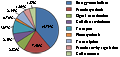 Figure 9 Biological functional classification of annotated genes |
2 Discussion
The grape berry, as the non-climacteric fruit, shows characteristic double-sigmoidal growth curve. The time of harvesting and the sugar/acid balance at the particular ripening stage affects the final quality of the resulting wine (Fillion et al., 1999). Therefore it is of the utmost importance to understand and characterize the berry ripening process, so as to control the maturation as much as possible. In this research, the ESTs analysis based on the cDNA library provided useful information about genes and functions expressed in the pericarp at veraison of developing berry. However, there is also 25.63% of the analyzed sequences which did not match any sequences in database after the sequencing analysis. Terrier et al (2001) found that the proportion of unmatched sequences was the highest at the veraison. Veraison is the onset of ripening, so some of these new genes encoding certain specific proteins could actually play an important role at the transient period which need further analysis to address their functions.
The striking changes in the mRNA population and gene expression occur in grape berries as the fruit enters into the ripening phase (Davies and Robinson, 2000). There is a range of genes whose transcription level increases at veraison, such as the genes encoding anthocyanin synthesis pathway enzymes (Boss et al., 1996). The anthocyanins, as the secondary metabolites localized in the skin of the berry, are responsible for quality factors, such as color and flavor. Among the 710 sequences analyzed, there were several genes related to the anthocyanin metabolism, and 32 ESTs in total were directly correlated with anthocyanidin synthesis, representing 3.3% of the total EST sequences. The structural genes involved with the process of anthocyanin synthesis included 4-coumarate-CoA ligase, chalcone synthase, chalcone isomerase, flavonoid 3',5'-hydroxylase, anthocyanidin 3-O-glucosyltransferase, glutathione S-transferase and so on. Other structural genes were not detected, which probably was due to the low abundance. The related genes of anthocyanin synthesis pathway, as a branch of flavonoid synthesis pathway, were highly expressed at the veraison (Table 4).
Table 4 BLAST matched correlative genes of biosynthetic pathway of anthocyanin in the GenBank |
The regulatory genes were first studied in model plants, such as maize, petunia and snapdragon, which were characterized in Arabidopsis (Davies and Schwinn, 2003; Broun, 2005). The main types of transcription factors that activate the structural genes and initiate the anthocyanin synthesis pathway effectively include MYB, MYC (bHLH), WD40, WKRY, Zinc finger and Homeodomain. In this pathway, the myb-related transcription factor was detected, and 12 clones representing 12 ESTs were relevant to zinc finger. The current mechanism of regulation accepted is that MYB and MYC interact as co-activators of the biosynthetic genes, and the WD40 repeat-proteins somehow assist in this process (Davies and Schwinn, 2003; Broun, 2005). And the tissue specificity of anthocyanin synthesis depends on this interaction. Furthermore, the control of the flavonoid pathway in different plant species is affected by the promoter regions of the anthocyanin structural genes, as well as the expression profiles and properties of the regulatory genes (Boss and Davies, 2009).
In wine grapes, acidity not only impacts the berry quality, but is also an important factor in deciding the harvest-date. And the sourness plays an important role in further processing of berries. Malate as the only high-proportion organic acid with active metabolism during ripening, is a must for the balance of acids in wine grape and favors desirable microorganisms growth for wine fermentation through preventing undesirable growth (Kunkee, 1991). Additonally, malate concentrationaffects secondary processes and thus the final characteristics of berries (grapes). There are two pathways of malate synthesis in fruit. The metabolism of sugars translocated to the berry enables synthesis of malate in the grape before veraison. Another means of malate synthesis in the berry is photosynthesis. After the veraison, malate starts to catabolize through various alleys, and it is likely a vital resource of carbon for certain pathways (Sweetman et al., 2009). Enzymes which are thought to be responsible for potential metabolic pathways involving malate in fruit cells were detected. These enzymesinclude alcohol dehydrogenase (ADH), isocitrate dehydrogenase (IDH), lactate dehydrogenase (LDH), pyruvate kinase (PK). ADH plays a central role in stress-response and could be a fruit-ripening related marker on grapevine. The up-regulation of some ADH genes for fruit-ripening implies that ethanol fermentation is a normal development response in grapes (Sarni-Manchado et al., 1997; Sweetman et al., 2009; Terrier et al., 2005). And Tesniere et al (2009) found that over-expression or down-regulation of VvADH resulted in unexpected responses at the primary and secondary metabolism levels in leaves and berries.
The study of grapevine disease tolerance and defense has entered the molecular era, which provides both a theory basis and the genetic tools to describe grapevine-pathogen interactions as well as discovering the resistance genes and other defense-related genes. PR-proteins are the particular class of proteins which are not expressed in plants without pathogen interaction or largely induced during infection (Gomès et al., 2009). The synthetic process of PR-proteins is one of the major steps in plant defense reaction, and the diversity of PR-proteins expressed decreases during ripening, which may be the reason for the enhanced susceptibility of the berries at the last stages of ripening (Monteiro et al., 2007).
The large-scale ESTs sequencing provided us useful information about genes and functions expressed in the pericarp at the veraison. To explore the precise functions of genes for understanding the biological characteristics of non-climacteric fruit, further analysis and research must be carried out. In 2007, the genome sequencing of V. vinifera L cv. Pinot Noir was completed, and the gene derived from the library constructed here contributes to the identification of the complete transcriptome of V. amurensis. The valuable source of cDNA clones can be helpful for gene mapping by PCR analysis and construction of high-density arrays for large-scale gene expression studies. Therefore, the development of the biochemical and genetic approaches can provide useful tools for getting more insight on the function of genes.
3 Materials and Methods
3.1 Plant material
V. amurensis (cv. Shuang Feng, 50% colored) berries were harvested during summer 2009 in National V. amurensis germplasm resource vineyards. Berries were frozen immediately in liquid nitrogen in the field and stored at -80℃. The pericarps were used as the experimental materials.
3.2 RNA extraction
Both cDNA library construction and generation of ESTs are RNA-based techniques. High quality RNA was isolated according to the optimized and modified CTAB protocol.
3.3 Preparation of cDNA library
Refer to the CreatorTM SMARTTM Library construction Kit User Manual for specific steps. 1 μL of total RNA was used to prepare the first-strand cDNA synthesis. 2 μL of the ss cDNA was used as the template for the long-distance PCR. The following primers were used for the PCR: 5'-AAGCAGTGGTATCAACGCAGAGTGGCCATTACGGCCGGG-3' (SMART Oligonucleotide), 5'-ATTCTAGAGGCCGAGGCGGCCGACATG-d(T)30N-1N-3' (CDS â…¢/3' PCR Primer) and 5'-AAGCAGTGGTATCAACGCAGAGT-3' (5' PCR Primer). A 5 μL sample of the PCR product was analyzed on a 1.1% agarose gel.
3 μL ds cDNA was proceeding to proteinase K treatment, then digested by Sfi I. The CHROMA SPIN-400 Column was prepared for cDNA size fractionation. And then the ds cDNA extracted and purified was ligated to the Sfi I-digested, dephosphorylated pDNR-LIB vector provided with the kit. The recombinant plasmid was transformed into E. coli DH5α by electroporation. The transformation was made up to 1 mL with LB broth and incubated with shaking for 1hr at 37℃.
3.4 cDNA library screening
The 50 μL dilution was spreaded onto a prewarmed 90 mm LB agar plate containing 30 μg/mL of chloramphenicol and the agar plate was incubated at 37℃ overnight. The plates were examined the next day. The desired transformation mixtures were pooled to generate the original, unamplified cDNA library.
20 isolated colonies were selected randomly to set up a PCR reaction using the M13 primers provided to screen the cDNA library. 5 μL of PCR product was electrophoresed on a 1.1% agarose gel with DNA size markers to determine the percentage of recombinant clones and the insert size. The titer was determined according to the CreatorTM SMARTTM Library construction Kit User Manual.
3.5 Large-scale EST sequencing and analysis
The BGI Limited was commissioned to be responsible for the assignment of cDNA library sequencing. The standard T7 sequencing primers which read into the presumed 3’ end of each cDNA were used in all sequencing reactions. Sequencing assignment was performed on the 3730xl DNA analyzer (Applied Biosystems). DNA sequence chromatograms were carried out using the phred software. Subsequently, sequences were vector-trimmed using Cross_Match software in the phrap package and quality-trimmed according to the quality value determined by phred. The final (processed?) sequences were submitted to the GenBank dbEST database. To identify unigenes, phrap was used to assemble ESTs into contigs with the parameters of 38 bp overlap length and 96% overlap identity.
3.6 Functional annotation of ESTs and unigenes
For annotation of the ESTs and unigenes, sequences were compared using BLASTX against both non-redundant protein database and non-redundant nucleic database at the NCBI site. Functional annotations were based upon searches against multiple databases in order to identify putative function of ESTs and unigenes. According to Gene ontology (GO) classification and BLASTX comparison, the sequence data were divided into different functional groups characterized according to putative and potential roles in berry development and compositions.
This study was supported by China Agriculture Research System (CARS-30).
Authors' Contributions
XNJ and BL planned and conducted experiments, analysed the data and wrote the first draft of the manuscript. WZ and CJY helped on a regular basis in data analysis. JW conceived the idea of the experiments and modified the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This study was supported by China Agriculture Research System (CARS-30).
References
Ablett E., Seaton G., Scott K., Shelton D., Graham M.W., Baverstock P., Lee L.S., and Henry R., 2000, Analysis of grape ESTs: global gene expression patterns in leaf and berry, Plant Science, 159(1): 87-95
http://dx.doi.org/10.1016/S0168-9452(00)00335-6
Boss P.K., and Davies C., 2009, Molecular biology of anthocyanin accumulation in grape berries, in the grapevine molecular physiology & biotechnology (Second Edition), Springer, Berlin, Germany, pp.263-292
Boss P.K., Davies C., and Robinson S.P., 1996, Analysis of the expression of anthocyanin pathway genes in developing V. vinifera L. cv Shiraz berries and the implications for pathway regulation, Plant Physiology, 111(4): 1059-1066
Broun P., 2005, Transcriptional control of flavonoid biosynthesis: a complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis, Current Opinion in Plant Biology, 8(3): 272-279
http://dx.doi.org/10.1016/j.pbi.2005.03.006 PMid:15860424
Coombe B.G., 1992, Research on development and ripening of the grape berry, American Journal of Enology and Viticulture, 43(1): 101-110
da Silva F.G., Iandolino A., Al-Kayal F., Bohlmann M.C., Cushman M.A., Lim H., Ergul A., Figueroa R., Kabuloglu E.K., Osborne C., Rowe J., Tattersall E., Leslie A., Xu J., Baek J.M., Cramer G.R., Cushman J.C., and Cook D.R., 2005, Characterizing the grape transcriptome, analysis of expressed sequence tags from multiple Vitis species and development of a compendium of gene expression during berry development, Plant Physiology, 139(2): 574-597
http://dx.doi.org/10.1104/pp.105.065748 PMid:16219919 PMCid:1255978
Davies C., and Robinson S., 2000, Differential screening indicates a dramatic change in mRNA profiles during grape berry ripening, cloning and characterization of cDNAs encoding putative cell wall and stress response proteins, Plant Physiology, 122(3): 803-812
http://dx.doi.org/10.1104/pp.122.3.803 PMid:10712544 PMCid:58916
Davies K.M., and Schwinn K.E., 2003, Transcriptional regulation of secondary metabolism, Functional Plant Biology, 30(9): 913-925
http://dx.doi.org/10.1071/FP03062
Fillion L., Ageorges A., Picaud S., Coutos-Thévenot P., Lemoine R., Charles Romieu C., and Delrotl S., 1999, Cloning and expression of a hexose transporter gene expressed during the ripening of grape berry, Plant Physiology, 120(4): 1083-1093
http://dx.doi.org/10.1104/pp.120.4.1083 PMid:10444092 PMCid:59342
Gomès E., and Coutos-Thévenot P., 2009, Molecular aspects of grapevine-pathogenic fungi interaction, in the grapevine molecular physiology & biotechnology (Second Edition), Springer, Berlin, Germany, pp.407-428
Ha D.T., Chen Q.C., Hung T.M., Youn U.J., Ngoc T.M., Thuong P.T., Kim H.J., Seong Y.H., Min B.S., and Bae K., 2009, Stilbenes and oligostilbenes from leaf and stem of Vitis amurensis and their cytotoxic activity, Archives of Pharmacal Research, 32(2): 177-183
http://dx.doi.org/10.1007/s12272-009-1132-2 PMid:19280145
Kunkee R.E., 1991, Some roles of malic acid in the malolactic fermentation in winemaking, FEMS Microbiology Leters, 88(1): 55-72
http://dx.doi.org/10.1016/0168-6445(91)90006-4 http://dx.doi.org/10.1016/0378-1097(91)90696-8 http://dx.doi.org/10.1111/j.1574-6968.1991.tb04957.x
Liu H.F., Yang C.J., Yu M., and Wang J., 2009, cDNA cloning and analysis of UDP- glucose: flavonoid 3-O-glucosyltransferase (3GT) in Vitis amurensis, Plant Physiology Communications, 45: 748-752
Liu H.F., Yang C.J., Zhao Q., Li B., and Wang J., 2009, cDNA cloning and analysis of flavonoid 3'-hydroxylase (F3'H) in Vitis amurensis Rupr, Plant Physiology Communications. 45: 1186-1190
Lund S.T., Peng F.Y., Nayar T., Reid K.E., and Schlosser J., 2008, Gene expression analyses in individual grape (Vitis vinifera L.) berries during ripening initiation reveal that pigmentation intensity is a valid indicator of development staging within the cluster, Plant Molecular Biology, 68(3): 301-315
http://dx.doi.org/10.1007/s11103-008-9371-z PMid:18642093
Monteiro S., Piçarra-Pereira M.A., Loureiro V.B., Teixeira A.R., and Ferreira R.B., 2007, The diversity of pathogenesis-reated proteins decreases during grape maturation, Phytochemistry, 68(4): 416-425
http://dx.doi.org/10.1016/j.phytochem.2006.11.014 PMid:17188723
Moser C., Segala C., Fontana P., Salakhudtinov I., Gatto P., Pindo M., Zyprian E., Toepfer R., Grando M.S., and Velasco R., 2005, Comparative analysis of expressed sequence tags from different organs of Vitis vinifera L, Functional & Integrative Genomics, 5(4): 208-217
http://dx.doi.org/10.1007/s10142-005-0143-4
Pacey-Miller T., Scott K., Ablett E., Tingey S., Ching A., and Henry R., 2003, Genes associated with the end of dormancy in grapes, Functional & Integrative Genomics, 3(4): 144-152
http://dx.doi.org/10.1007/s10142-003-0094-6
Peng F.Y., Reid K.E., Liao N., Schlosser J., Lijavetzky D., Holt R., Zapater J.M.M., Jones S., Marra M., Bohlmann J., and Lund S.T., 2007, Generation of ESTs in Vitis vinifera wine grape (Cabernet Sauvignon) and table grape (Muscat Hamburg) and discovery of new candidate genes with potential roles in berry development, Gene, 402(1-2): 40-50
http://dx.doi.org/10.1016/j.gene.2007.07.016 PMid:17761391
Sarni-Manchado P., Verriès C., and Tesnière C., 1997, Molecular characterization and structural analysis of one alcohol dehydrogenase gene (GV-Adh1) expressed during ripening of grapevine (Vitis vinifera L.) berry, Plant Science, 125(2): 177-187
http://dx.doi.org/10.1016/S0168-9452(97)04630-X
Sweetman C., Deluc L.G., Cramer G.R., Ford C.M., and Soole K.L., 2009, Regulation of malate metabolism in grape berry and other developing fruits, Phytochemistry, 70(11-12): 1329-1344
http://dx.doi.org/10.1016/j.phytochem.2009.08.006 PMid:19762054
Terrier N., Ageorges A., Abbal P., and Romieu C., 2001, Generation of ESTs from grape berry at various development stages, Journal of Plant Physiology, 158(12): 1575-1583
http://dx.doi.org/10.1078/0176-1617-00566
Terrier N., Glissant D., Grimplet J., Barrieu F., Abbal P., Couture C., Ageorges A., Atanassova R., Léon C., Renaudin J-P., Dédaldéchamp F., Romieu C., Delrot S., and Hamdi S., 2005, Isogene specific oligo arrays reveal multifaceted changes in gene expression during grape berry (Vitis vinifera L.) development, Planta, 222(5): 832-847
http://dx.doi.org/10.1007/s00425-005-0017-y PMid:16151847
Tesniere C., and Abbal P., 2009, Alcohol dehydrogenase genes and proteins in grapevine, in the grapevine molecular physiology & biotechnology (Second Edition), Springer, Berlin, Germany, pp.141-161
Weidner S., Karamać M., Amarowicz R., Szypulska E., and Gołgowska A., 2007, Changes in composition of phenolic compounds and antioxidant properties of Vitis amurensis seeds germinated under osmotic stress, Acta Physiologiae Plantarum, 29(3): 283-290
http://dx.doi.org/10.1007/s11738-007-0035-4
. PDF(955KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Xiangnan Ji
. Bo Li
. Wen Zhang
. Chengjun Yang
. Jun Wang
Related articles
. Wine grape ( Vitis amurensis )
. cDNA library
. Unigene annotation
. Gene ontology
. EST sequencing
Tools
. Email to a friend
. Post a comment


