Research Article
Genetic Assessment of Amaranthus Linn. Genotypes in Treatment Combinations of Glomus clarum and Leucaena leucocephala Lam. Using Simple Sequence Repeat (SSR) Marker 
 Author
Author  Correspondence author
Correspondence author
Molecular Plant Breeding, 2017, Vol. 8, No. 10 doi: 10.5376/mpb.2017.08.0010
Received: 14 Jan., 2017 Accepted: 26 Mar., 2017 Published: 15 Sep., 2017
Olawuyi O.J., and Onuoha S.O., 2017, Genetic assessment of Amaranthus Linn. genotypes in treatment combinations of Glomus clarum and Leucaena leucocephala Lam. using simple sequence repeat (SSR) marker, Molecular Plant Breeding, 8(10): 85-99 (doi: 10.5376/mpb.2017.08.0010)
The molecular and genetic assessments of the seeds of five Amaranthus genotypes in four treatment combinations of Arbuscular mycorrhiza Fungus (Glomus clarum) and Green manure (Leucaena leucocephala) were evaluated using SSR maker with four primers. The experiment was laid out in a complete randomized design with four replicates. Heritability of growth traits were higher than yield traits for treated Amaranthus genotypes. The plant height had strong positive correlation with stem length (r=0.8919), inflorescent length (r=0.6150) and inflorescent width (r=0.6004). Prin 1 accounted for the highest variation with proportion of 0.4197 and eigenvalue of 5.8765. The study detected the polymorphism, gene and allelic diversity and established phylogenetic relationships among the Amaranthus population. NGBO1271 treated with G. clarum + L. leucocephala had the highest concentration of extracted DNA (4039.2ul), while NGBO1234 treated with G. clarum and NGBO1234 treated with L. leucocephala had highest genomic DNA nanodrop of 2.14 gl. A total of 15 alleles were detected and number of allele per primer ranged from 3 to 5 with mean value of 3.75. The highest allelic frequency of 0.75 was recorded for ASAAC005 and ASAAC006. However, ASAAC001 primer was polymorphic (66.86%), with highest allele number and allele diversity of 5.0000 and 0.7200 respectively. The result from Nei’s coefficient also confirmed genetic variation based on the total gene diversity and gene diversity per locus. The dendogram among the treated Amaranthus genotypes showed their phylogenetic relationship which can be useful in selection of closely related genotypes for desirable traits. Therefore, NGBO1234 and NGBO1271, G. clarum and L. leucocephala could be suitable genotypes and further explored in genetic improvement of crops. Also, ASAAC001 primer could be considered in molecular breeding of Amaranthus and other vegetables.
Background
Amaranthus is a green vegetable that belongs to the family Amarantheceae (Tucker, 1986; Bressani et al., 1992). It survives under harsh conditions, and is of economic, nutritional and medicinal values (Breene, 1991; Gupta and Gudu, 1991; Asfaw, 1997). Thus, the improvement of Amaranthus may be useful for alleviating hunger in developing nations, especially in overpopulated and undernourished areas (Pal and Khoshoo, 1974; Sauer, 1993).
The vegetable helps to build immunity in human, and ensure proper functionality of organs and tissues through provision of vitamins, minerals like calcium, iron and phosphorous and other phytochemicals (George, 2003; Nnamani et al., 2007). The fiber content also prevents constipation but consumed as a staple food during ancient times (Noonan, 1999; Buragohain et al., 2013). Amaranthus species such as A. blitus, A. cruentus, A. hypochondriacus are often planted for leaves, whereas A. caudatus, A. hypochodriacus, A. cruentus, A. hybridus are planted for their grain (Caselato-Sousa and Amaya-Farfn, 2012).
Amaranthus species consist of grain and weedy types, with the common grain types being A. hypochodriacus, A. cruentus, A. caudatus and the major weedy types include; A. viridis, A. spinosus, A. retroflexus and A. hybridus.
Arbuscular Mycorrhiza Fungi (AMF) plays an important role in the uptake of water, nutrients and improves crop performance and soil quality (Koske and Polson, 1984; Osonubi et al., 1995; Neveen et al., 2008). They help plants to capture nutrients such as phosphorus, sulphur, nitrogen and micronutrients from the soil (Brundrette, 2002). Glomus clarum is one of the Arbuscular Mycorrhiza Fungi (AMF) that supports the growth and improves yields of plants (Olawuyi et al., 2012; Fapohunda et al., 2013).
Simple Sequence Repeat (SSR) is PCR-based marker with the highest information content when evaluating and characterizing the germplasm (Powell et al., 1996). They are reliable, reproducible, cheap and discriminative compared to other markers (Smith et al., 1997). Simple Sequence Repeat (SSR) works on the principle of microsatellite assay. It anchors based on the fact that, it is highly polymorphic even between closely related lines. It requires low amount of DNA and can be easily automated for high throughput screening which can be exchanged between laboratories but are highly transferable between populations (Gupta et al., 1999). SSRs are mostly co-dominant markers for studies on population genetics and mapping of genes (Jarne and Lagoda, 1996; Goldstein and Schlotterer, 1999). The use of fluorescent primers in combination with automatic capillary or gel-based DNA sequencers had been adopted in most advanced laboratories, while SSR are excellent markers for fluorescent techniques, multiplexing and high throughput analysis.
Despite the global food insecurity, there is need to provide information on suitable SSR primer and bio inoculant to establish genomic relationship among Amaranthus genotypes. This will enhance the genetic management and conservation of Amaranthus.
This study investigated the molecular and phylogenetic relationship among Amaranthus germplasm in treatment combinations of Arbuscular Mycorrhiza Fungi (AMF) and Leucaena leucocephala (GM) using Simple Sequence Repeat (SSR) marker.
1 Results
The nanodrop and DNA concentration of the extracted Amaranthus genotypes were found to be at 260/280/gl in Table 1. The gel results showing the PCR Amplification of DNA in Amaranthus genotypes is presented in Figure 1. The Polyacrylide Gel Electrophoresis (PAGE) of treated Amaranthus genotypes using primers; ASAAC001, ASAAC005, ASAAC006 and ASAAC011 are shown in Figure 2, Figure 3, Figure 4 and Figure 5 respectively.
|
Table 1 Nanodrop and DNA concentration of Amaranthus genotypes in treatment combinations of AMF (Glomus clarum) and GM (Leucaena leucocephala) Note: G1 = NGB01271; G2 = NGB01644; G3 = NGB01234; G4 = NGB01613; G5 = NGB01662; T1 = Control; T2 = AMF (Glomus clarum) only; T3 = GM only (Leucaena leucocephala); T4 = AMF + GM (Glomus clarum + Leucaena leucocephala) |
|
Figure 1 PCR Amplification of genomic DNA of Amaranthus genotypes in treatment combinations of Glomus clarum and Leucaena leucocephala |
|
Figure 2 PAGE of 20 Amaranthus genotypes obtained using AHAAC001 |
|
Figure 3 PAGE of 20 Amaranthus genotypes obtained using AHAAC005 |
|
Figure 4 PAGE of 20 Amaranthus genotypes obtained using AHAAC006 |
|
Figure 5 PAGE of 20 Amaranthus genotypes obtained using AHAAC011 |
The growth and yield of Amaranthus treated with GM (Leucaena leucocephala) are shown in Figure 6 and Figure 7. The quality of genomic DNA concentration was generally good with the highest total concentration of genomic DNA of 4039.20 ul recorded for combinations of Glomus clarum and Leucaena leucocephala on NGBO1271. The Nanodrop of genomic DNA of G. clarum for genotype NGBO1662 had the highest value of 2.17 gl followed by NGBO1234 for G. clarum (2.14 gl) and NGBO1662 for L. leucocephala (2.14 gl).
|
Figure 6 Growth of Amaranthus cultivars treated with GM |
|
Figure 7 The yield of Amaranthus cultivars treated with GM |
A total of four primers of SSR marker revealed the diversity and major allele frequency as well as the polymorphic information content of five Amaranthus genotypes in treatment combinations of G. clarum and L. leucocephala (Table 2). The result showed that primer ASAAC001 was highly polymorphic (66.86%) with the highest allele number and allele diversity of 5 and 0.7200 respectively compared with other primers. ASAAC005 and ASAAC006 primers had the highest major allele frequency of 0.7500. Four (4) amplified microsatellite loci were revealed by ASAAC001 and ASAAC005 primers while three (3) were produced by ASAAC006 and ASAAC011 primers.
|
Table 2 Frequency and diversity of allele and polymorphic information content (PIC) of treated Amaranthus Genotypes using SSR marker |
The result of Nei’s Coefficient of gene variation (GST), gene diversity per locus and total gene diversity are shown in Table 3. The mean total for Nei’s Coefficient of gene variation (GST), gene diversity per locus and total gene diversity were 0.2275, 0.3391, and 0.4389 respectively.
|
Table 3 Nei's analysis of gene variation and gene diversity per locus in Amaranthus population Note: Ht: Total gene diversity; Hs: Gene diversity per locus; Gst: Nei's coefficient of gene variation or measure of genetic differentiation |
The dendogram showing the phylogenetic relationships of Amaranthus genotypes in treatment combinations of G. clarum and L. leucocephala is shown in Figure 1. There are two major clusters sub-divided into six (6) groups in which clusters 4 and 6 had the highest number of genotypic treatment while cluster 1 had least with only NGBO1662 of G. clarum and L. leucocephala combinations. Glomus clarum treated with NGBO1271 is not closely related to untreated NGBO1644, NGBO1271 treated with G. clarum and L. leucocephala, untreated NGBO1271 and L. leucocephala treated with NGBO1271 in sub-cluster 6. G3T3, G3T1 and G3T2 are genetically related than G4T1 and G3T4 in cluster 4. G5T4 and G5T3 are closely related than G5T1 and G4T2, G5T2 in cluster 3 and G4T3 are genetically similar in cluster 2. Again, G2T2 and G2T3 are related compared to G2T4 (Figure 8)
|
Figure 8 Dendrogram showing the genetic relationships of the genotypes of treated Amaranthus spp Note: G1 = NGB01271; G2 = NGB01644; G3 = NGB01234; G4 = NGB01613; G5 = NGB01662; T1 = Control; T2 = AMF only; T3 = GM only; (Leucaena leucocephala); T4 = AMF + GM |
The result of the mean performance for genotypic effect of growth related characters for treated Amaranthus reveals significant (P < 0.01) effect on Amaranthus genotypes as shown in Table 4. NGB01662 was significantly higher for plant height compared to other genotypes. While, number of leaves, stem length, stem girth, leaf length and leaf width were significantly influenced in NGB01271. Also, the plant height of NGBO1644, and NGBO1613 as well as NGBO1234, NGBO1644, NGBO1613 and NGBO1662 for number of leaves were not significantly different from one another.
|
Table 4 Genotypic effect on growth performance of Amaranthus in treatments combinations of Glomus clarum (AMF) and Leucaena leucocephala (GM) Note: Mean with the same letter in the same column is not significantly at P ≥ 0.05 according to Duncan Multiple Range Test (DMRT) |
The result of the mean performance for genotypic effect of yield related characters for Amaranthus reveals significant (P<0.01) effect on Amaranthus genotypes (Table 5). NGB01662 was significantly higher for inflorescence length and inflorescence width compared to other genotypes. Also, the numbers of inflorescence were significantly higher for NGB01234 but different from other genotypes. NGB01271 is significantly higher for wet yield of inflorescent, dried yield of inflorescent, wet leaf and dried leaf than other genotype while number of branches was significantly higher in NGB01613.
|
Table 5 Genotypic effect on yield characters of Amaranthus treated with Glomus clarum and Leucaena leucocephela Note: Means with the same letter in the same column are not significantly at P ≥ 0.05 according to Duncan Multiple Range Test (DMRT) |
The plant height, stem length, and leaf width was significantly (p<0.05) increased for Leucaena leucocephala (Green Manure) than other treatments and control (Table 6).
|
Table 6 Effect of AMF, GM and combinations of AMF + GM on growth characters of Amaranthus Note: Mean with the same letter in the same column are not significantly at P≥0.05 according to Duncan Multiple Range Test (DMRT); AMF = Glomus clarum; GM = Leucaena leucocephala |
The result in Table 7 shows that Glomus clarum, Arbuscular Mycorrhiza Fungus (AMF) is significantly higher for Inflorescence length while Leucaena leucocephala (Green Manure) is significantly higher for Inflorescence width, wet yield of inflorescent, dried yield of inflorescent and dried leaf. Also, the combined treatments of AMF + GM is significantly higher for number of inflorescence and wet leaf, while untreated plant was higher for number of branches.
|
Table 7 Effect of AMF, GM and their combinations on yield characters of Amaranthus Note: Mean with the same letter in the same column are not significantly at P ≥ 0.05 according to Duncan Multiple Range Test (DMRT); AMF = Glomus clarum; GM = Leucaena leucocephala |
The component of variance for growth and yield traits in treated Amaranthus genotypes shown in Table 8 indicates that the phenotypic variance of both growth and yield characters were higher than the genotypic variance in all the characters. The values for the phenotypic and genotypic variances were highest for number of leaves while the least were recorded for stem girth and inflorescence width. The heritability was highest for dried yield of inflorescent (0.739) while the least was recorded for inflorescence width (-0.022).
|
Table 8 Heritability, phenotypic and genotypic variance of growth and yield characters in treated Amaranthus genotype |
The result from Table 9 delineates the Amaranthus genotype into fourteen principal component axes (Prin. 1-14). Prin. 1 which constituted the highest accounted for 0.4197 of the proportion eigenvalue of 5.876478, while Prin. 14 was the least with proportion of 0.0006 and eigenvalue of 0.008592. The result in Prin 1 shows that plant height, stem length, leaf breadth, leaf length, inflorescent length are closely related to one another compared to inflorescent breadth and number of inflorescent which are related to each other.
|
Table 9 Principal components analysis (PCA) of growth and yield characters of treated Amaranthus genotypes Note: PH: Plant height; NL: Number of leaves; SL: Stem length; SG: Stem girth; LL: Leaf length; LB: Leaf width; NI: Number of inflorescent; IL: Inflorescent length; IB: Inflorescent width; NB: Number of branches |
The correlation result in Table 10 showed that plant height was positive and strongly correlated with stem length (r=0.89 p<0.01), inflorescent length (r=0.62), inflorescent width (r=0.60) and a positive correlation with leaf width (r=0.54). Number of leaf had a strong significant positive correlation with stem girth (r=0.68), leaf length (r=0.61), number of branches (r=0.77), wet leaf (r=0.61) and a positive correlation with dried leaf (r=0.57). stem length had a positive significant correlation with stem girth (r=0.64), leaf width (r=0.65) and dried leaf (r=0.60) but a positive correlation with leaf length (r=0.52) and wet leaf (r=0.59). Stem girth is positive and strongly associated with leaf length (r=0.75), leaf width (r=0.64), number of branches (r=0.69), wet leaf (r=0.77) and dried leaf (r=0.73). Leaf length is also positive and related with leaf breadth (r=0.67) and wet leaf (r=0.60) but a positive correlation with number of branches (r=0.58), and dried leaf (r=0.59). Leaf width had a significant positive correlation with wet leaf (r=0.57) and dried leaf (0.55). Inflorescent length had a strong significant positive correlation with inflorescent width (r=0.91) and number of inflorescent (r=0.73). Inflorescent width had a strong significant positive correlation with number of inflorescent (r=0.78). Wet yield of inflorescent had a strong significant positive correlation with dried yield of inflorescent (r=0.69).
|
Table 10 Correlation co-efficient among characters in treatments of AMF and GM in Amaranthus genotype Note: PH: Plant height; NL: Number of leaves; SL: Stem length; SG: Stem girth; LL: Leaf length; LB: Leaf Width; NI: Number of inflorescent; IL: Inflorescent Llength; IB: Inflorescent width; NB: Number of branches |
2 Discussions
Genetic assessment and improvements in Amaranthus germplasm in treatment combinations will play a key role in future studies and improvements of vegetable crop.
G1T4 had highest concentration of extracted DNA while G3T2 had highest genomic DNA nanodrop. This implies that good quality extracted DNA is enhanced when Amaranthus is treated with combinations of AMF and GM.
When treated singly with Glomus clarum and Leucaena leucocephala and a combination of AMF and GM, NGB01662 produced high mean for Inflorescence length and inflorescence width compared to other genotypes. Also, NGB01234 produced high mean for Number of inflorescence compared to other genotypes. NGB01271 produced high mean for wet yield of inflorescent, dried yield of inflorescent, wet leaf and dried leaf than other genotype while NGB01613 produced high mean for number of branches. Moreover, the treatments had a very high mean for the number of inflorescence, inflorescence length, inflorescence breadth, number of branches, fresh leaf biomass, and dried leaf biomass, wet yield of inflorescent and dried yield of inflorescent.
Also, NGB01662, NGB01644 and NGB01234 produced a higher mean for plant height and stem length compared to other genotypes. plant height, number of leaves, stem length, stem girth, leaf length and leaf breadth produced high mean for NGB01271 while NGB01613 produced high mean for plant height, number of leaves, stem length and stem girth. However, NGB01271 performed best for growth and yield in treatment combinations of AMF and GM. This supported the findings made by Olawuyi et al., (2012) and Babajide et al., (2012).
AMF produced high mean for inflorescence length. GM produced high mean for Inflorescence width, wet yield of inflorescent, dried yield of inflorescent and dried leaf. AMF+GM produced high mean for number of inflorescence and wet leaf while CONTROL produced high mean for number of branches. The performance of GM implies that it can be harnessed for agricultural productivity in order to reduce the effect of chemical fertilizers as reported by Sobulo (2000).
Notable amongst the molecular marker used for genetic diversity studies, linkage map construction, and marker-assisted selection (MAS) is Microsatellites (Chung et al., 2009; Zhao et al., 2011). All primers used were polymorphic across all the Amaranthus genotype with amplified microsatellites loci. This shows a further indication of the ability of these primer pairs to distinguish different genotypes of Amaranthus in treatment combinations.
There were variations in major allele frequency, number of amplified microsatellite loci, number of allele and allele diversity. This supported the report of Mallory et al., (2008). Mandal and Das (2002) observed a high degree of genetic diversity when they worked with grain Amaranthus using RAPD markers, which was similar to the report of Transue et al. (1994) and Chan and Sun (1997). However, Plaschke et al. (1995) confirmed that a relatively small number of SSRs are sufficient to discriminate closely related bread wheat cultivars. These studies revealed that high levels of genetic differences and similarities can still exist amongst the Amaranthus genotypes even in treatment combinations.
As co-dominant and locus specific markers, SSR primer pairs can be used to monitor line uniformity between and within lines of a clone. This information can assist breeders in selecting the appropriate treatment to be applied on Amaranthus so as to obtain maximum yield and equally maintain genetic variability. The SSR markers were found to be suitable for genetic mapping studies, which will assist efforts towards marker assisted selection (MAS) in Amaranthus. The results from this study also clearly demonstrated the effectiveness of the SSR markers for fingerprinting Amaranthus spp. genotype as a means of agronomic improvement.
Molecular markers allowed us to estimate the overall genetic similarities and differences in Amaranthus and simultaneously to reveal molecular-based genetic relationships. Cluster analysis and dendogram, indicates that cluster groups consist of genotypes from different treatment combinations. The cluster diagram among the treated Amaranthus genotype showed that there were genetic similarities and differences. Thus, high inter- and intraspecies variability exists even among genotypes (Mosyakin and Robertson, 1996; Štefúnová et al., 2014). Moreover, genotypes with different treatments were found in different clusters. Such wide adaptability has been attributed to similarity in selection history, requirements, cultivations and developmental traits. The relationships that exist among the genotypes in the clusters show that there were genetic similarities which were similarly reported by Bamgbegbin et al. (2016). Identification and conservation of germplasm are necessary for maintaining genetic diversity, studying local genetic material in order to choose ecotypes having high nutritional interest in their place of origin (Perez-Gonzalez, 2001). Moreover, the genetic information obtained from this study could aid the design of useful and appropriate tool for selecting representative genotypes and effectively managing Amaranthus germplasm breeding programs.
3 Conclusions and Recommendation
ASAAC001 primer detected the highest polymorphism, allele and gene diversity compared to other primers. This suggests that this primer could be used for further molecular breeding of other vegetables. Since Amaranthus has been confirmed as an under-utilized crop, this will prevent its gradual loss of genetic diversity.
The establishment of genetic similarity and phylogenetic relationship among Amaranthus genotypes in treatment combinations of bioinoculants could further be an efficient means of introducing novel variability into Amaranthus gene pool. NGB01271 genotype could be selected, while GM, AMF and combinations of AMF and GM could be recommended for crop improvement breeding programs with a view to adopt Integrated Nutrient Management Approach (INMA) for yield improvement and proper documentation of germplasm conservation of Amaranthus spp for future diversity studies.
4 Materials and Methods
4.1 Germplasm collections, study location and plant samples
The seeds of the five genotypes of Amaranthus spp. evaluated in this research study were collected from the National Centre for Genetic Resource and Biotechnology (NACGRAB), Moor Plantation, Ibadan, Nigeria. The genotypes evaluated were; NGB01271, NGB01644, NGB01234, NGB01613 and NGB01662. The molecular studies were carried out in the Laboratory of Bioscience Unit of International Institute of Tropical Agriculture (IITA), Ibadan, Nigeria. Fresh leaves samples from the five genotypes of Amaranthus were collected very early in the morning by 8:00 am after two weeks of planting. The collected leaf samples were kept in a cold environment at a temperature of 0˚C so as to prevent denaturation of their DNA contents. A total of five leaf samples were randomly collected per treatment from the three replicates for DNA extraction. Twenty leaf samples were collected altogether from each block. From the whole experimental set-up, a total of one hundred leaf samples were collected altogether and lyophilized at -80˚C.
4.2 Planting procedure
The field Experiment was a complete Randomized Design which was properly spaced at 22 cm between treatment and 65 cm between blocks. The genotypes were subjected to treatments as follow, T1 = uninoculated (control), T2 = 5 g AMF and T3 = 5 g GM while T4 had a combination of 2.5 g AMF and 2.5 g GM. replicated thrice. The treatments comprised of Arbuscular Mycorrhiza Fungus (AMF), Green Manure (Leucaena leucocephala) and combinations of Arbuscular Mycorrhiza Fungus (Glomus clarum) and Green Manure (Leucaena leucocephala). These treatments were applied after two weeks of transplanting. The set-ups were moderately watered to resist drought and enhance development.
4.3 DNA extraction and quantification
Extraction of DNA from fresh leaf sample was the first step using molecular marker and primers. The DNA was extracted from fresh and lyophilized young leaf samples of Amaranthus in order to obtain good quality DNA. This DNA extraction was carried out according to Dellaporta et al., (1983) protocol modified in sodium dodecyl sulphate (SDS) extraction buffer. One gram of fresh young leaves of Amaranthus was grinded using mortar and pestle followed by the addition of one litre of extraction buffer. The extraction buffer consisted of isopropanol, 10 mg/ml RNASE A, β-mercaptoethanol, 1 % PVP (1 g of 100 ml), 5 M NaCl (292.2 g in 1, 000 L), 1 M Tris (121.4 in 1,000 ml, PH 8), 0.5M EDTA (146.12 g in 1,000 L, PH8), 20% SDS (200 g SDS + 800 ml of double distilled water), 70% ethanol (70 ml of ethanol in 100 ml of double distilled water), 25 ml CIA (24 ml chloroform + 1 ml Isoamyl Alcohol), 5 M Potassium Acetate (490.7 in 1,000 ml) stirred at 4˚C, Low salt TE buffer (10 ml 1 M Tris HCL, 2 ml EDTA, 950 ml of double distilled water and adjusted to PH to 8).
One litre of this buffer was prepared using, 100 ml 1 M Tris HCL + 50 ml 0.5M EDTA + 100 ml 5 M NaCl + 1% PVP.
DNA extraction protocols involved grinding or digesting cellular constituents in order to release the content. Detergents such as sodium dodecyl sulphate (SDS), Cetyl trimethylammonium bromide (CTAB) was used for the removal of membranes lipids. When the DNA is released, it is protected from endogenous nucleases by the inclusion of EDTA in the extraction buffer which is necessity for the chelating magnesium ions that are a significant co-factor for nucleases. The eppendorf tubes were thoroughly mixed and incubated at 65˚C for 60 minutes in a water bath. 5 ml of 5 M potassium acetate (PH 5.5) was added to each of the tubes and kept for 20 minutes at 4˚C in ice. The mixture was centrifuge at 15,000 rpm for 15 minutes and 10 ml of ice cold isopropanol was added to the supernatant and kept for incubation at 4˚C for 30 minutes. The solutions were centrifuged at 15,000 rpm for 15 minutes and the pellets were dissolved in sterile double distilled water. The DNA extract usually contains sizeable amount of RNA, proteins, polysaccharides, tannins and pigments that may interfere with the extracted DNA. An addition of protein degrading enzymes known as proteinase-K is use to remove the proteins, followed by denaturation at 65˚C and precipitation using chloroform and Isoamyl alcohol. Also, RNAs are normally removed using RNA degrading enzyme known as RNase A. The DNA solutions was transferred to 2 ml eppendorf tube and treated with RNAses (10 mg/ml) for 1 hr at 37˚C and 1 ml of chloroform:isoamyl alcohol (24:1) was added and Centrifuged at 12,000 rpm for 15 minutes. Though, Polysaccharide-like contaminants are, more tasking to remove. However, the combinations of NaCl and CTAB have been observed to remove polysaccharides (Murray and Thompson, 1980; Paterson et al., 1993). Moreover, some protocols replace NaCl with KCl (Thompson and Henry, 1995). Since DNA will be released along with other compounds like lipids, proteins, carbohydrates or phenols. It needs to be separated from the other compounds by centrifugation. The DNA in the aqueous phase will then be transferred into new eppendorf tubes without disturbing the interphase and ice cold ethanol was added to precipitated the DNA in salt solution (e.g. sodium acetate) or alcohol (100% isopropanol or ethanol), re-dissolved in sterile water or buffer. The precipitate was centrifuged at 12,000 rpm for 10 minutes and the supernatant discarded. The pellet was washed in 70% ethanol; air dried and finally dissolved 100 ul of sterile double distilled water. The determination of DNA concentration extracted needs to be measured using 1% agarose gel electrophoresis and detected using UV illuminator or spectrophotometer. Agarose gel checks whether the DNA is degraded or not but estimating DNA concentration by visually comparing band intensities of the extracted DNA with a molecular ladder of known concentration is too subjective. Spectrophotometer measures the intensity of absorbance of DNA solution at 260 nm wavelength, and also indicates the presence of protein contaminants but does not determine whether the DNA was degraded or not.
The DNA quantification was done to ascertain the quality and quantity of DNA extracted from the plant samples. Purity of the DNA in the samples, dissolved in TE buffer was analyzed by checking the absorbance ratios at 260/280 nm on nanodrop spectrophotometer followed by determination of concentration.
4.4 Primers design of simple sequence repeat (SSR) marker
Primers flanking each unique microsatellite were designed using the web-based computer program Primer 3 version 2.0 (Rozen and Skaletsky, 2000) according to the program’s default parameters, with the following exceptions: preferred product size range equal to 150-200 base pairs; melting temperature differences in forward and reverse primers of no more than 1˚C; and max poly-X (maximum allowable length of a mononucleotide repeat) of three. These primers were designed and used for the flanking regions of the SSRs by Melanie Ann Mallory in 2007. Oligonucleotide primers were synthesized by Inquaba Biotech, Ibadan. Primer pairs were assigned names based on their repeat motif that is AS = Amaranthus spp, AAC = motif type while clone ID were 001, 005, 006 and 011 which resulted to a total of four primers having both forward and reverse sequence were selected from the designed by Melanie Ann Mallory (2007) (Table 11).
|
Table 11 The primers, their motifs and sequences |
4.5 Polymerase chain reaction (PCR), amplification conditions and amplification of DNA product
PCR reactions for SSRs was carried out in the presence of forward and reverse primers that anneal at the 5` and 3` ends of the template DNA respectively. PCR fragments are usually separated on polyacrylamide gels in combination with AgNO3 staining, autoradiography or fluorescent detection systems. Agarose gels (usually 3%) with EtBr can also be used when differences in allele size among samples is larger than 10 bp.
The amplification conditions was: an initial step of denaturation for 5 minute at 94˚C followed by 44 cycles each consisting of a denaturation step of 15 seconds at 94˚C, an annealing step of 20 seconds at 65˚C and an extension step of 30 seconds at 72˚C. Seven minutes will be given after the last cycle to the extension step at 72˚C to ensure the completion of the primer extension reaction followed by hold temperature at 10˚C lasting for infinity. The PCR products were separated on 5% polyacrylamide gel at 80 volt, 300 mA 60Watt for 1 hour 30mins. It was then visualized and photographed using silver staining under UV transilluminator (Zhang et al., 2002).
Amplification of microsatellite loci were carried out in 10 μl PCR reactions containing 3.0 ul of 100 ng/ul total genomic DNA, 0.5 μl of 5pMol of forward and reverse primers each, 0.8 ul DMSO, 0.4 ul of 50 mM MgCl2, 0.8 ul of 2.5 Mm DNTPs, 2.9 ul H2O and 0.1 ul of Taq DNA polymerase using Master Mix. Amplification was performed using an eppendorf thermocycler.
4.6 Polyacrylamide gel electrophoresis (PAGE)
The reagents used was prepared from a mixture of the following; 40 ml of water, 25 ml of TBE, 7.5 ml of polyacrylamine, 50 µl of Terred and 500 µl of APS was added last.
The procedures included the glass comb and rubbers were washed in order to dry. The glass plate and spacer were wiped with 70% ethanol. The glass plate was lined with rubber and the spacer was placed between both spacer. The plates were carefully clipped together (the blue clip is used to clip the plates together, while the white clip is used to clip the electrophoretic tank.
When the glass tank was well arranged and positioned, the mixed reagent was poured into it until it gets to the brim. It is allowed for some minutes to gel. When it gels, it is then inserted into the gel tray where the DNA samples are introduced using a staining dyes alongside the 1 kb plus ladder gene ruler from Thermos scientific for formation of bands. The gel was then brought out after 1 hour 30 mins and loaded into the chamber for viewing using the UV trans illuminator.
4.7 Determination of growth and yield characters
Data taken on morphological characters included plant height (cm), number of leaves, stem length (cm), stem girth (cm), and leaf area (cm2). At maturity period, data were also collected on the quantitative and qualitative flower characters which included; number of inflorescence, inflorescence width (cm), inflorescence length (cm) and number of branches. The Genotypic variance, phenotypic variance and Heritability were determined using the following formula:
.jpg)
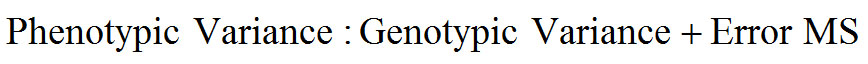
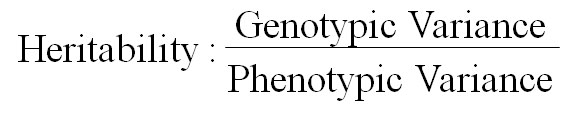
4.8 Statistical analysis
The data on growth and yield characters were subjected to Analysis of Variance (ANOVA) using SAS 9.3 software while the means were separated by DMRT.
Molecular data generated was subjected to molecular analysis in order to generate information on Total gene diversity, Gene diversity per locus using PopGene Version 1.32 (Yeh and Boyle, 1999) and Powermarker V3.25 (Liu and Muse, 2005). Nei's coefficient of gene variation and genetic distance was calculated according to Nei (1972). These values were used to generate dendogram using Unweighted Pair Group Method with Arithmetic Average (UPGMA) cluster analysis as described by Sneath and Sokal (1973) to reveal phenetic representations of genetic relationship among the treated Amaranthus spp.
Asfaw Z., 1997, Conservation and use of traditional vegetables in Ethiopia. In: L Guarino (ed). International.Workshop on Genetic Resources of Traditional Vegetables in Africa.Institute of Plant Genetics and Crop Plant Research Rome, J Pl Food Hum Nut, 48 (3): 57-65
Bamigbegbin B. J., Olawuyi O. J., and Jonathan S. G., 2016, Molecular variability of Celosia argentea using amplified fragment length polymorphism (AFLP) marker, Molecular Plant Breeding, 7(26): 1-6
Breene W.M., 1991, Food uses of grain amaranth, Cereal Foods World, 36:426–430
Bressani R., 1989, The proteins of grain amaranth, Food Rev Int., 5:13–38
https://doi.org/10.1080/87559128909540843
Bressani R., Gonzalez J.M., Zuniga J., Bruener M., Elias L.G., 1987, Yield, selected chemical composition and nutritive value of 14 selections of amaranth grain representing four species, J Sci Food Agric, 38:347–356
https://doi.org/10.1002/jsfa.2740380407
Bressani R., Sanchez-Marroquin A., Morales E., 1992, Chemical composition of grain amaranth cultivars and effects of processing on their nutritional quality, Food Rev Int, 8:23–49
https://doi.org/10.1080/87559129209540928
Brewbaker J.L., and Sorensson C.T., 1990, New tree crops from interspecific Leucaena hybrids, In: Janick, J. and Simon, J.E (eds) Advances in New crops, pp. 283-289
Brundrett M. C., 2002, Coevolution of roots and mycorrhizas of land plants. New Phytologist 154, 275–304
https://doi.org/10.1046/j.1469-8137.2002.00397.x
Buragohain J., Singh V.B., Deka B. C., Jha A. K., Wanshnong K., Angami T., 2013, Collection and evaluation of some underutilized leafy vegetables of Meghalaya, Indian Journal of Hill Farming, 26(2):111-115
Caselato-Sousa V.M., and Amaya-Farfn J., 2012, State of knowledge on amaranth grain: A comprehensive Review, Journal of Food Science, 77:93-104
https://doi.org/10.1111/j.1750-3841.2012.02645.x
Chung H.K., Kim K.W., Chung J.W., Lee J.R., Lee S.Y., Dixit A., Kang H.K., Zhao W.G., McNally K.L., Hamilton R.S., Gwag J.G., and Park Y.J., 2009, Development of a core set from a large rice collection using a modified heuristic algorithm to retain maximum diversity, Journal of Integrative Plant Biology, 51:1116-1125
https://doi.org/10.1111/j.1744-7909.2009.00882.x
Dellaporta S.L., Wood J., Hicks J.B., 1983, A plant DNA minipreparation: version II. Plant Mol. Biol. Rep., 1:19-21
https://doi.org/10.1007/BF02712670
Fapohunda S.O., Olawuyi O.J., and Okei C.P., 2011, Antimicrobial and phytochemical potentials of arbuscular mycorrhizal fungi in Nigeria, The South Pacific Journal of Natural and Applied Sciences, 29 : 21-25
Fapohunda S.O., Olawuyi O.J., Bello O.B., and Lawal T., 2013, Comparative Shoot Responses of two Nigerian Crops to Glomus clarum and other Fertilizers, Greener Journal of Agricultural Sciences, 3 (4):280-285
https://doi.org/10.15580/GJAS.2013.3.021913478
George P.M., 2003, Encyclopedia of foods.Volume 1, Humane Press, Washington, pp. 526
Goldstein D.B., Schlotterer C., 1999, Microsatellites, evolution and applications, Oxford University Press, New York
Gupta P.K., Varshney R.K., Sharma P.C., Ramesh B., 1999, Molecular markers and their applications in wheat breeding, Plant Breed, 118:369–390
https://doi.org/10.1046/j.1439-0523.1999.00401.x
Gupta P.K., Rustgi S., Kulwal P.L., 2005, Linkage disequilibrium and association studies in higher plants: Present status and future prospects, Plant Mol Biol, 57: 461–485
https://doi.org/10.1007/s11103-005-0257-z
Gupta V.K., and Gudu S., 1991, Interspecific hybrids and possible phylogenetic relations in grain amaranths, Euphytica, 52: 33-38
Jarne P., and Lagoda P., 1996, Microsatellites, from molecules to populations and back, Trends Ecol. Evol. 11: 424–429
https://doi.org/10.1016/0169-5347(96)10049-5
Liu K., Muse S.V., 2005, PowerMarker: an integrated analysis environment for genetic marker analysis, Bioinformatics, 21: 2128–2129
https://doi.org/10.1093/bioinformatics/bti282
Liu Z.W., Biyashev R.M., Saghai Maroof M.A., 1996, Development of simple sequence repeat DNA markers and their integration into a barley linkage map, Theor Appl Genet, 93 : 869-876
https://doi.org/10.1007/BF00224088
Liu Z.W., Jarret R.L., Kresovich S., Duncan R.R., 1995, Characterization and analysis of simple sequence repeat (SSR) loci in seashore paspalum (Paspalum vaginatum Swartz.), Theor Appl Genet, 91:47-52
https://doi.org/10.1007/BF00220857
Mallory M.A., Hall R.V., McNabb A.R., Pratt D.B., Jellen E.N., Maughan P.J., 2008, Development and characterization of microsatellite markers for the grain amaranths, Crop Sci, 48: 1098–1106
https://doi.org/10.2135/cropsci2007.08.0457
Mosyakin S.L., and Robertson K.R., 1996, New infrageneric taxa and combination in Amaranthus (Amaranthaceae), Ann.Bot. Fenn., 33: 275-281
Mosyakin S.L., Robertson K.R., 2003, Amaranthus. In: Flora of North America. North of Mexico. Oxford University Press, New York, USA
Murray M.G., Thompson W.F., 1980, Rapid isolation of high molecular weight plant DNA, Nucleic Acids Res., 8:4321-4325
https://doi.org/10.1093/nar/8.19.4321
Nei M., 1972, Genetic distance between populations. Am. Nat. 106: 283–292
https://doi.org/10.1086/282771
Nnamani C.V., Oselebe H.O., Okporie E.O., 2007, Ethnobotany of Indigenous Leafy Vegetables of Izzi Clan, in Ebonyi State, Nigeria. In: Proceeding of 20th Annual National Conference of Biotechnology Society of Nigeria. Abakaliki, November 14th -17th, pp.111-114
Noonan S.C., Savage G.P., 1999, Oxalate content of foods and Its effect on humans, Asia Pactific J. Clin. Nutr., 67: 64-74
Oboh B., 2007, Multivariate analysis of the diversity among some Nigerian accessions of Amaranthus hybridus, Int. J. Plant Breed. Genet., 1: 89-94
https://doi.org/10.3923/ijpbg.2007.89.94
Olawuyi O.J., Babatunde F.E., Akinbode O.A., Odebode A.C., and Olakojo S.A., 2011, Influence of Arbuscular mycorrhizal and N.P.K fertilizer on the productivity of cucumber (Cucumis sativus), International Journal of Organic Agriculture Research and Development, 3 : 22-31
Olawuyi O. J., Ezekiel-Adewoyin D. T., Odebode A. C., Aina D. A. and Esenbamen G. E., 2011, Effect of arbuscular mycorrhizal (Glomus clarum) and organo-mineral fertilizer on growth and yield performance of okra (Abelmoschus esculentus), African Journal of Plant Science, 6(2): 84-88
Osonubi O., Atayese M.O., and Mulongoy K., 1995, The effect of vesicular-arbuscular mycorrhizal inoculation on nutrient uptake and yield of alley-cropped cassava in a degraded Alfisol of southwestern Nigeria, Biology and Fertility of Soils, 20: 70-76
https://doi.org/10.1007/BF00307844
Pal M., Khoshoo T.N., 1974, Grain amaranths. In: Hutchinson JB (ed) Evolutionary studies in world crops: diversity and change in the Indian subcontinent. Cambridge University Press, UK, pp.129-137
Pal M., Ohri D., and Subrahmanyam., 2000, A new basic chromosome number for Amaranthus (Amaranthaceae). Cytologia, 65: 13-16
https://doi.org/10.1508/cytologia.65.13
Palaniappan S. P., Jeyabal A., and Chelliah S., 1999, Evaluation of Integrated Nutrient Management in Summer Sesame (Sesamum indicum L.), Sesame and Safflower Newsletter, No. 14
Paterson A.H., Brubaker C.L., Wendel J.F., 1993, A rapid method for extraction of Cotton (Gossypium spp) genomic DNA suitable for RFLP or PCR analysis, Plant Mol. Biol. Rep., 11:122-127
https://doi.org/10.1007/BF02670470
Perez-Gonzalez S., 2001, The importance of germplasm preservation and use for temperate zone fruit production in the tropics and subtropics. In: Perez-Gonzalez S, Dennis F, Mondragon C, Byrne D, editors. VI International symposium on temperate fruit growing in the tropics and subtropics, Mexico: Acta Hort (ISHS), 565: 25–32
Powell W., Morgante M., Andre C., Hanafey M., Vogel J., Tingey S. and Rafalski A., 1996, The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis, Molecular Breeding, 2: 225-238
https://doi.org/10.1007/BF00564200
Ray T., and S. Ch. Roy., 2009, Genetic diversity of Amaranthus species from the Indo-Gangetic plains revealed by RAPDanalysis leading to the development of ecotype-specific SCAR marker, J. Heredity, 100(3): 338-347
https://doi.org/10.1093/jhered/esn102
Rozen S., and Skaletsky H. J., 2000, Primer3 on the WWW for general users and for biologist programmers. In Krawetz, S. and Misener, S. (eds) Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press, Totowa, NJ, pp.365-386
http://www-genome.wi.mit.edu/genome_software/other/primer3.html
Sauer J.D., 1950, The grain amaranths and their relatives: a survey of their history and classification, Ann Mo Bot Gdn., 37:561–619
https://doi.org/10.2307/2394403
Sauer J.D., 1967, The grain amaranths and their relatives: a revised taxonomic and geographic survey, Annals of the Missouri Botanical Garden, 54: 103-137
https://doi.org/10.2307/2394998
Sauer J.D., 1976, Grain amaranths. In: Simmonds NW (ed) Evolution of crop plants. Longman, London, pp.4–7
Sauer J.D., 1993, Amaranthaceae—amaranth family. In: Historical Geography of Crop Plants: A Select Roster. CRC, Boca Raton, Florida, USA, pp.9–14
Smith D.N., Devey M.E., 1994, Occurrence and inheritance of microsatellites in Pinus radiata, Genome, 37: 977-983
https://doi.org/10.1139/g94-138
Smith J.S.C., Chin E. C.L, Shu H., Smith O.S., Wall S.J., Senior M.L., Mitchell S.E., Kresovich S., and Ziegle J., 1997, An evaluation of the utility of SSR loci as molecular markers in maize (Zea mays. L.), comparison with data from RFLPs and pedigree, Theorectical Applied Genetics, 95: 163-173
https://doi.org/10.1007/s001220050544
Smith S.E., Read D.J., 1997, Vesicular Arbuscular mycorrhiza in agriculture and horticulture. In: mycorrhiza symbiosis. Second edition, Smith, SE, DJ Read (eds), Academic Press, London, UK, pp.453-469
Sneath P. H. A., and Sokal R. R., 1973, Numerical taxonomy — The principles and practice of numerical classification. W. H. Freeman, San Francisco, pp.573
Snezana Drinic Mladenovic, Kostadinovic Marija, Ristic Danijela, Simic Milena, Stefanovic Lidija. 2012, Assessment of genetic relatedness of the two Amaranthus retroflexus populations by protein and random amplified polymorphic DNA (RAPD) markers, Afr J Biotechnol., 11(29): 7331–7337
Štefúnová V., Bežo M., Labajová M., and Senková S., 2014, Genetic analysis of three Amaranth species using ISSR markers, Emir. J. Food Agric., 26(1): 35-43
Štefúnová V., Bežo M, Ziarovská J., and Ražná K., 2015, Detection of the genetic variability of Amaranthus by RAPD and ISSR markers pak, J. Bot., 47(4): 1293-1301
Sun M., Chen H., and Leung F.C., 1999, Low-Cot DNA sequences for fingerprinting analysis of germplasm diversity and relationships in Amaranthus, Theor. Appl.Genet., 99: 464-472
https://doi.org/10.1007/s001220051258
Thompson D., Henry R.J., 1995, Single step protocol for preparation of plant tissue for analysis by PCR, Biotechniques, 19:394-397
Tucker J.B., 1986, Amaranth: The once and future crop, Bioscience, 36:9-13
https://doi.org/10.2307/1309789
Xu F.X., and Sun M., 2001, Comparative analysis of phylogenetic relationships of grain amaranths and their wild relatives (Amaranthus; Amaranthaceae) using internal transcribed spacer, amplified fragment length polymorphism, and double-primer fluorescent intersimple sequence repeat markers, Molecular Phylogenetics and Evolution, 21(3): 372-387
https://doi.org/10.1006/mpev.2001.1016
Yeh F.C., and Yang R., 1999, Microsoft window-based freeware for population genetic analysis (POPGENE Ver. 1.31), Canada, AB: University of Alberta
Zhao W.G., Chung J.W., Cho Y.I., Rha W.H., Lee G.A., Ma K.H., Han S.H., Bang K.H., Park C.B., Kim S.M., and Park Y.J., 2010, Molecular genetic diversity and population structure in Lycium accessions using SSR markers, Comptes Rendus Biologies, 333:793-800
Zhao W.G., Chung J.W., Lee G.A., Ma K.H., Kim H.H., Kim K.T., Chung I.M., Lee J.K., Kim N.S., Kim S.M., and Park Y.J., 2011, Molecular genetic diversity and population structure of a selected core set in garlic and its relatives using novel SSRmarkers, Plant Breeding, 130:46-54
https://doi.org/10.1111/j.1439-0523.2010.01805.x
. PDF(1233KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Odunayo Joseph Olawuyi
. S.O. Onuoha
Related articles
. Amaranthus
. Glomus clarum
. Leucaena leucocephala
. SSR primer
Tools
. Email to a friend
. Post a comment
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)


