Over Expression of Chitinase and Chitosanase Genes from Trichoderma harzianum under Constitutive and Inducible Promoters in order to Increase Disease Resistance in Wheat (Triticum aestivum L) 
2. Department of Molecular Phytopathology and Genetics, University of Hamburg, Ohnhorststr. 18, D-22609 Hamburg, Germany
3. Centre of Agricultural Biochemistry and Biotechnology, University of Agriculture, Faisalabad-38040-Pakistan
 Author
Author  Correspondence author
Correspondence author
Molecular Plant Breeding, 2012, Vol. 3, No. 4 doi: 10.5376/mpb.2012.03.0004
Received: 05 Mar., 2012 Accepted: 16 Apr., 2012 Published: 30 Apr., 2012
Rana et al., 2012, Over Expression of Chitinase and Chitosanase Genes from Trichoderma harzianum under Constitutive and Inducible Promoters in order to Increase Disease Resistance in Wheat (Triticum aestivum L), Molecular Plant Breeding, Vol.3, No.4 37-49 (doi: 10.5376/mpb.2012.03.0004)
Powdery mildew of wheat, caused by Erysiphe graminis f.sp. tritici is a serious threat to wheat grain production in many parts of the world. Efforts have been made to engineer resistance into wheat using single gene technology which culminated in the improved resistance efficiency but never a complete control. Some efforts of expressing genes for disease resistance under the control of constitutive promoter resulted in the production of abnormal plants. We used double gene technology and over expressed a couple of antifungal genes i.e., chitinase and chitosanase from Trichoderma harzianum simultaneously into winter wheat genotype Florida. They were expressed separately under the constitutive promoter (Ubiquitin-1) from Maize and stress cum disease inducible promoter (Vst-1) from Vitis vinefera. Six lines achieved under constitutive promoter and five lines under inducible promoter showed the co-integration and expression of chitinase and chitosanase genes. All these lines showed multiple copy integration of both these genes except one line which showed single copy integration of chitinase under ubiquitin promoter. The copy number varied for Chitinase (1-3) and Chitosanase (2-10). Different promoters did not seem to have any impact on transformation efficiency. Pathological analysis done with E.graminis showed a decrease in the susceptibility for both types of transgenic plants. A decrease in susceptibility was seen upto 75% when the transgenics were under inducible promoter. While in transgenics under constitutive promoter the decrease in susceptibility of upto 60% was seen. All the primary transforments with the exception of a couple showed normal growth.
Introduction
Wheat is one of the most important food crops of the world. It is grown almost throughout the world, is the most grown crop and ranks second after Maize in terms of production. It is the staple food in many parts of the world and is rich source of proteins, carbohydrates and vitamins. Wheat is consumed throughout the world as an important part of the diet, i.e., in the form of bread, noodles, spaghetti, cakes, biscuits and other sweets in Europe and America, while in the form of chapatti in Asia and Africa. In Indo-Pak subcontinent wheat is the most popular staple food and is the symbol of food security (Hammad et al., 2011, Khan and Khan, 2010). Wheat is attacked by many pathogens including fungi, viruses, bacteria and nematodes but the fungal diseases are the most common. Rusts, smuts, powdery mildews and scab are of global importance and have caused from little damage to 70% yield losses (Naz et al., 2007).
Plant diseases can be controlled by cultural practices, biological control, use of fungitoxic chemicals and/or by the development of resistant genotypes. Only the resistant genotype is the stable and risk free method to control plant diseases. However the resistance sources are limited in wheat. Genetic engineering provides a source for interphyletic gene transfer to create new resistance sources. Wheat transformation was started in the early 1990’s. Since then many reports have emerged on the optimization, improvement of protocols and use of these protocols for the transfer of genes of agronomic and economic importance. Many attempts have been done to engineer fungal resistance in wheat using single gene transfer technology which has brought partial resistance against pathogens under question. The strategy has been to target fungal cell wall or fungal metabolism by using the genes encoding proteins which either digest and degrade fungal cell wall or interfere metabolic processes, the examples has been glucanases, chitinases, thaumatin like protein and ribosomal inactivating proteins (Muehlbauer and Bushnell, 2003, Bliffeld et al., 1999 and Bieri et al., 2003, Oldach et al., 2001, Chen et al., 1999; Makandar et al., 2006; Balconi et al., 2007; Mackintosh et al., 2007; Shin et al., 2008).
Structurally fungal cell wall is consisted of β-glucan and amino glucans chitin (Ply-GluNAc), some portion of which is always acetylated in the form of chitosan. The attempts till present have targeted either β-glucan with the help of glucanase gene or chitin with the help of chitinase (Oldach et al., 2001; Mackintosh et al., 2007). In both the cases single genes are unable to completely disrupt the fungal cell wall, additionally the chitosan is never degraded by chitinase and this portion helps the fungus to survive. A successful strategy will be to target both the chitin and chitosan by overexpressing chitinase and chitosanase genes. By this strategy not only both amino glucans will be targeted but the impact of the degradative enzymes will be boosted synergistically resulting in completely dismantled cell wall structure.
Constitutive expression of transgene is generally desirable but in disease resistance related experiments it has not proved as rule. Sometimes the transgenes have shown disease like symptoms under normal conditions when the pathogen attack was not there. This is probably due to cellular reprogramming in the presence of proteins which are desired only under specific conditions. Similarly the examples have proved that constitutive promoters are not necessarily constitutive all the time and there is different expression in different parts of the plant. Under such conditions it was not possible to get the disease control in those plant parts where constitutive promoter did not express at desired level. Inducible promoter is the solution of such pitfalls.
1 Results
With the emergence and development of molecular biology, the phenomenon of disease resistance and pathogen virulence is being studied at molecular level. The factors responsible for pathogen virulence and plant defense responses against pathogen attacks are being studied extensively. Parasites of pathogens are being used as tools for biological control of plant diseases. All these attributes of plants and the pathogens are controlled by genes and the manipulation of these genes by genetic transformation can be used for increasing resistance of plants against diseases. In this research, two antifungal genes HarChit and HarCho from Trichoderma harzianum (myco-parasite) were co-expressed separately under constitutive Ubiquitin promoter as well as under stress cum disease inducible Vst-1 promoter in wheat. These genes hydrolyse the chitin present in the fungal cell wall. Inducible and constitutive promoters were used to get advantages of both the promoters and compare the effect on the disease resistance. To evaluate any of their effect on disease resistance wheat fungal pathogen Erysiphe graminis f. sp. tritici, the causal organism of powdery mildew of wheat was used in infection assays using transgenic lines and non transgenic controls.
1.1 Genetic transformation of wheat
Winter wheat genotype Florida was selected for transformation purpose. This genotype has already been reported to be readily transformable by many authors. Immature Zygotic Embryos were bombarded in a batch of pUbi-HarChit, pUb-HarCho and P35SAcS and then separately making a batch of pVst-HarChit, pVst-HarCho and P35SAcS. P35SAcS is a selection marker gene which confers resistance against Basta. The bombarded IZEs were maintained on a regime of callus induction and regeneration media containing selection agent Basta. The results are summarised in Table 1. The plants recovered upon selection media were checked by PCR and Southern Blot analysis for the presence or absence of genes of interest. All the plants recovered through selection did not contain genes of interest rather only 8 out of 34 Basta selected plants contained at least one gene of interest in case of Experiment-1 (pUbi-HarChit+Ubi-HarCho+P35SAcS) and 6 out of 31 plants contained at least one gene of interest in case of Experiment-2 (pVst-HarChit+Vst-HarCho+P35SAcS). This made a transformation frequency of 0.26 and 0.17% for first and second combination. If talked only based upon Basta resistance which is conferred by P35SAcS, the transformation frequency proved to be 1.08 and 0.87% for both combinations as a total of 3128 and 3540 IZEs were bombarded respectively.
Table 1 Primer pairs used to amplify HarChit and HarCho gene |
1.2 Molecular verification of pUbi-HarChit and pUbi-HarCho transgenic plants and their progenies
The plants which were resistant to Basta were checked for the presence of pUbi-HarChit and/or pUbi-HarCho by Southern Blot analysis. Details of the procedure can be seen in the material and Methods. A 400 bp long fragment from HarChit gene and another 400 bp long fragment from HarCho gene were labelled with DIG. Chemiluminescence’s detection was done with CSPD® and the images were developed on X-rays film. A total of seven plants (I.A-1, I.A-2, I.A-3, I.A-4, I.A-4, I.A-5, I.A-6, and I.A-18) had pUbi-HarChit construct with intact promoter, gene of interest and terminator. Out of these seven plants three had single copy of pUbi-HarChit, three had double copy integration and one had three to four copies integrated in the genome. Seven plants (I.A-1, I.A-3, I.A-4, I.A-4, I.A-5, I.A-6, I.A-17 and I.A-18) were positive for pUbi-HarCho and out of these seven plants six were already positive for pUbi-HarChit. I.A-2 was positive for pUbi-HarChit but proved negative for pUbi-HarCho. Similarly I.A-17 was negative for pUbi-Harchit but proved positive for pUbi-HarCho. Out of the seven plants positive for the pUbi-HarCho three showed single copy integration, one showed two copies, others showed four, five and seven copy integration. In summery six plants showed the co-integration of both pUbi-HarChit and pUbi-HarCho constructs while single construct (once pUbi-HarChit and once pUbi-HarCho) was present in couple of plants. The rest of the plants were either having only bar gene or were wrongly selected.
1.3 Molecular verification of pVst-HarChit and pVst-HarCho transgenic plants
The results of Southern Blot analysis showed that out of 31 plants only five were positive for both pVst-HarChit and pVst-HarCho while one plant was having only pVst-HarCho. The rest of the plants were either having only bar gene or were false selection. Only one plant had single copy integration for pVst-HarChit, the rest of the plants were having multicopy integrations for both the gene constructs. The copy number of integration varied from 1-5 for pVst-HarChit and 2-7 for pVst-HarCho. In the plant “I.A-8” one band in the lane reserved for cassette out showed heavier band than the expected while the bands in the lane reserved for linearized plasmid were normal. The rest of the plants showed at least one band equal to the band in the positive control lanes. The figure 1 shows the Southern Blot analysis for pVst-HarCho construct of all the positive plants.
 Figure 1 Southern blot analysis of primary transforments (T0). Note: U = Lane undigested g-DNA, 1 = Lane with g-DNA digested with endo-nuclease that linearizes the gene construct. In this case it is Kpn-I, 2 = g-DNA digested with endonuclease that cuts the cassette out of the gene construct. In this case it is sfi-I . Lanes1-3 = I.A-7, Lanes 4-6 = I.A-8, Lanes 7-9 = I.A-9, Lanes 10-12 = I.A-10, Lanes 13-15 = I.A-11, Lanes 16-18 = I.A-12, Lanes 19-23 = controls and empty lanes in between. |
DNA of all the probes along with positive and negative probes were run on 0.8% agarose gel and transferred to nylon membrane by capillary method and cross linked later. DNA on this membrane was hybridized to the DIG labelled 400 bp single stranded DNA probe from HarCho and the detection was made with CSPD substrate. Southern Blot analysis of the putative transgenic plants showed that 6 plants are positive for pVst-HarCho. Five out of them were also positive for pVst-HarChit while I.A-8 was negative for pVst-HarChit. They showed high copy number in general except for I.A-8 who had 1-2 copies.
1.4 Molecular verification of transgenes in segregating generation
Self pollinated T0 plants were harvested after they reached the maturity. Segregation analysis based on the BASTA resistance was done before going for molecular verification of the segregating generations. In all the lines except one from the first batch 3:1 ratio in the surviving to dead was seen while from the second batch of transgenics only two lines showed normal 3:1 ratios and the others did not follow the trend. It means single locus integration for bar gene was there in some lines and absent in others. The matter was not studied further in detail because it was beyond the scope of this project. The plants were advanced further for molecular analysis of genes of interest and phyto-pathological studies.
Selected Basta resistant plants from the transgene progenies were analyzed for the presence and cosegregation of HarChit and HarCho genes with bar gene. For both the batches with constitutive and inducible promoter respectively, HarChit and HarCho was always present in Basta resistant plants with the same trend and integration pattern seen in the primary transgenic. We have a case where all the plants died upon basta spray in the progeny but southern showed that HarCho and HarChit was still present in these plants. Progenies were not only checked in T1 but also in T2 example of one southern was shown in the figure 2.
Figure 2 Southern blot analysis I.A-11in T1 for Inducible promoter construct with HarChit gene specific probe |
DNA of all the probes along with positive and negative probes were run on 0.8% agarose gel and transferred to nylon membrane by capillary method and cross linked later. DNA on this membrane was hybridized to the DIG labelled 400 bp single stranded DNA probe from HarCho and the detection was made with CSPD substrate. Southern Blot analysis of the T1 selected plants showed that the integration of pVst-HarChit in the next generation is the same as it was in the primary transforments. T0 plant and the T1 selected plants looked the same with a copy number of three proving the successful transfer of pVst-HarChit.
1.5 Expression analysis of pUbi-HarChit and pUbi-HarCho transgenes by Northern Blot
In the first batch of transgenics HarChit and HarCho were under the control of constitutive promoter. Therefore all the plants which were proved to have pUbi-Harchit and/or pUbi-HarCho by Southern Blot analysis were checked for the expression of HarChit and HarCho genes in primary transforments as well as in T1 generation. RNA was isolated from the leaves of the transgenic and non transgenic control plants and run on the degrading agarose gel at the rate of 15 µg per lane per plant. Five lanes were reserved for each transgenic plant, one lane for T0 RNA and four for T1 only one lane was reserved for non transgenic control. RNA was transferred to the positively charged Nylon membrane. 400 bp long gene specific radio labelled DNA probes generated using primer pairs given in table 2 were used to detect the mRNA on memberane. Hybridization detection was done by capturing the illumination on X-Rays film. Experiment example is shown in figure 3. All the plants that contained HarChit and/or HarCho also showed expression of these genes though in varying levels, except I.A-5 that did not show expression of HarChit and HarCho although both of them showed their appearance when checked through Southern.
 Figure 3 Northern analyses of T0 and T1 generation with pUbi-HarChit construct Note: Freshly isolated RNA from young spikes of the entire Southern Blot analysis positive T0 and selected T1 progeny plants was run on 1% degeneration gel at the rate of 15 µg per probe/lane (3A) and transferred to the Nylon membrane by capillary method under clean environment and cross linked later. This RNA was then hybridized with 400 bp long radio actively labelled HarChit specific single stranded DNA and were detected on X-Rays film. Here I.A-18, I.A-5, I.A-2 and I.A-17 are transgenic lines. –ve= is the RNA from control non transgenic plant. T0 is the RNA from primary transgenic plant and T1P1 to T1P4 is the RNA from four selected plants from progeny of the primary transforments. |
Table 2 Primer sequences used for Southern, Northern and rtPCR |
1.6 Induction of HarChit and HarCho genes under the control of Vst1 promoter
The expression of HarChit and HarCho genes was induced by wounding with sea sand on the leaf surfaces in the transgenics where they were under Vst1 promoter. RNA was isolated eight hours after induction as reported by Leckband, 1997 and cDNA was synthesized. Later on PCR reactions were done using c-DNA as template and gene specific primers for both HarChit and HarCho genes. All the five plants which were Southern Blot analysis positive for pVst-HarChit showed an expected band of 400 bp and all the six plants positive for pVst-HarCho showed an expression for HarCho gene. The PCR product from all of them was compared with negative (non transgenic) control, PCR (H2O was used as second negative to check any contamination) and positive control (plasmid DNA specific product). The PCR products were also sequenced to confirm the HarChit and HarCho induction in transgenics. So we had co-expression of HarChit and HarCho genes in 5 transgenic lines and expression of only HarCho in one line (Figure 4).
 Figure 4 Induction of HarChit under stress inducible promoter in T0 and T1 generations Note: cDNA was synthesized from total RNA isolated from T0 and T1 plants and the amplification was taken using gene specific primers. Positive control showed the band of the expected size as did all the positive plants while the negative controls never showed the presence of any transcript from HarChit. PCR reaction showed the presence of HarChit transcript after Vst1 promoter induction with wounding stress. 4A= T0 plants and 4B= plants from T1 generation. M= Marker, C1= reagents control with H2O as template, C2 = negative control with template from non transgenic plant. C3 = positive control with template from pVst-Chit plasmid. |
1.7 Phyto-pathological experiments
Erysiphe graminis f.sp. tritici was used to check the effect of HarChit and HarCho genes expressed constitutively and induced under pathogen stress for the enhancement of plant response against diseases.
1.8 Transgenic lines under constitutive promoter
Leaf segments of 4 centimetres length were cut from plantlets at two leaves stage and cultured on the anti-seniscence media. Non transgenic control was compared with all the transgenic lines under study as well as with the transgenic control. It was made sure that 300-400 conidia should spread per cm2 of the inoculation plate. The results showed that there was almost no difference in the number of colonies developing on transgenic plants, transgenic control or negative control in the beginning. The difference started to be prominent 10 days post inoculation. The colonies remained healthy on the transgenic control and negative control while started withering and burning on the transgenic plants. The difference in the size was clearly visible three weeks post inoculation. This size was noted and compared with transgenic and non transgenic control. The relative number of colonies compared to size of colonies developed of non transgenic control is shown in Figure 5 and Figure 6. There was 34 to 60 percent less fungal growth on tested transgenic lines compared to control lines. I.A-18 showed 0.4 and I.A-6 showed 0.66 colony size compared to 1.0 of control, the other transgenic lines performed in between. Even the line I.A-1 that expressed only HarChit showed reduction. It also seems from these experiments that there is no role of bar or gus genes for fungal resistance.
The transgenics having HarChit and HarCho under the control of constitutive promoter showed reduction in fungal growth in general while the transgenics where HarChit and HarCho were under inducible promoter, 3 out of six tested plants showed no affect on fungal growth while in other three, the colony size of 0.74 and 0.64 was noted for I.A-7 and I.A-12 in comparison to 1.0 of the controls. I.A-11 proved to be the best in the lot and showed a colony size of 0.25 in comparison to 1 of the control. I.A-11 was the most resistant against Erysiphe among all the transgenic lines whether expressing antifungal genes under constitutive or inducible promoter. Even the number of developing colonies those were not less in number in transgenic lines and the control plants were on average 30% less than in number on I.A-11. Each experiment for pathological analysis was repeated at least thrice.
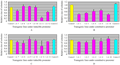 Figure 5 Detached leaf assay was done by spraying the small leaf segments with fungal powdery mass containing mycelium and spores. The results showed that the number of developing colonies on the transgenics under constitutive (C) and inducible control (D) decreased in some transgenic lines but the difference only slight with only one exception (IA-11) where ≥ 30% less colonies were detected. When we saw the colony health and measured the size of colonies randomly 21 dpi. Colony size was smaller and the fungal mass was dead in general on transgenic lines under constitutive promoter (B) while in the set of transgenic plants under inducible promoter the colony size did not change in some cases and was at par with control lines while in other cases it changed (A). The best reduction was shown by IA-11 which showed 75% smaller size compared to controls. Control= Non transgenic Florida leaf segments, Control2= transgenic Florida but containing bar and gus genes, not HarChit/HarCho |
Figure 6 Comparison of leaf segments inoculated with E. graminis 21dpi. Leaf segments on antiseniscent media without inoculations (C) remained healthy without any mycelia development while inoculated leaves always showed mycelia development; it was lesser in the form of small and dried colonies on transgnic leaf segments, healthy and large colonies on non transgenic leaf segments (A &B) |
2 Discussions
2.1 Wheat transformation, transformation frequency and segregation in the progenies
The objective of present study was the stable transformation of wheat with antifungal genes from Trichoderma harzianum. This was to be done separately under constitutive and inducible promoters so that the difference in the performance of both type of expression could be noted. Wheat transformation via biolistic bombardment is being reported in many important experiments of developmental and applied biology of wheat (Becker et al., 1994, Sivamani et al., 2000, Oldach et al., 2001, Anand et al., 2003, Becker et al., 2007 and Shin et al., 2008). In all these studies wheat transformation frequency has been reported nearly 1% of the total explant tissues tried. There is only one report where Pellegrineschi et al., 2002 reported the transformation frequency of more than 60% in 8 sister lines of genotype ‘Bobwhite‘. Our results did not match with Pellegrineschi et al., 2002 rather with others. Primary transgenics showed single copy to multicopy integration for both the genes of interest under inducible and constitutive promoters. There was only one plant where there was both the genes of interest present under constitutive promoter but their expression could not be detected using Northen Blot analysis. Similarly one line was seen where selection marker gene was completely silent in T1 generation while genes of interest were present expressed under inducible promoter. Recently Anand et al., 2003, reported that 20 out of 24 wheat transgenic plants transformed with rice chitinase and glucanse genes using pat as marker gene showed complete gene silencing in T1 generation and onwards. They observed heavy methylation of their transgene. Silensing of marker gene in our results may also be following the same phenomenon. We also noted an additional thing that many of the putative transgenic plants resistant to selectable marker were not containing our gene(s) of interest. Similarly there were instances when only one gene of interest was present in a transgenic instead of both. Transformation experiments were done over a year but the transgenic plants were found between November and April, this was what found by Brettschneider et al., 1997 during Maize transformation under Hamburg conditions. This indicates that there is something related with the explants which comes from outer environment of green house and affects its efficiency for transformation. The primary transgenics having the genes of interest were grown to maturity and the harvested seed was grown to get T1 generation where seven plants out eight for experiment-1 (genes of interest under constitutive promoter) showed single locus integration while only two out of six for could show the single locus integration for experiment-2 (genes of interest under constitutive promoter). This might be due to silencing of selection marker gene or any unknown reason.
2.2 Co-expression of HarChit and HarCho
The defence response genes function in a variety of ways to inhibit fungal infection and expression of these genes in transgenic plants has been shown to enhance fungal resistance (Muehlbauer and Bushnell, 2003). Wheat like other plants has an innate defence response against the fungal pathogens that involves the induction of PR genes at the rear end. These PR genes defend by attacking different organs of the pathogen. For example chitinases and 1, 3-β-glucanases hydrolyse the fungal cell wall by targetting fungal cell wall chitin (poly-GlcNAc) and 1, 3-β-Glucan as substrate. The presence of chitinase genes in plants and lower organisms have been known and the chitinolytic activity is also established invivo and invitro for these genes (Yanai et al., 1992; Blaisean and Lafay, 1992; Botha et al., 1998, Singh et al., 2007). Chitosanase is an enzyme similar to chitinase, capable of hydrolysing the -1,4-linkages between N-acetyl-D-glucosamine and D-glucosamine residues in a partially acetylated fungal cell wall polymer. Chitosanases have the potential of slowing or preventing fungal infection by degrading the structural chitosan (poly-GlcN) found in the cell wall of many fungi (Hendrix and Stewart, 2002). Glucose amine oligomers, released from fungal cell walls after hydrolysis with chitinase or chitosanase, are elicitors of plant defence response (Lee et al., 1999; Vander et al., 1998). The response elicited by these molecules depends on the length and degree of acetylation of the oligomers released (Vander et al., 1998). Oligomers that are relatively short (e.g. products of chitosanase hydrolysis) are active elicitors of plant defence systems.
T. harzianum, a soil-borne fungus known to be a control agent of fungal plant pathogens (Papavizas, 1985), is one of the fungi that produce degrading enzymes which destroy key cell wall structural polymers of fungal pathogens (Hendrix and Stewart, 2002). The purified enzymes from T. harzianum are substantially more chitinolytic and glucanolytic than the enzymes from other known sources (upto 100 times more active than the corresponding plant enzymes and effective on a much wider range of pathogens) (Lorito et al., 1994; Lorito et al., 1996). They are also non toxic to plants even at high concentration (Carsolio et al., 1999). Furthermore, the anti-fungal activity is synergistically enhanced when different Trichoderma cell wall degrading enzymes are used together or in combination with plant PR-proteins, commercial fungicides, cell membrane-affecting toxins or biocontrol bacteria (Lorito et al., 1996; Steyaert et al., 2004). Co-expression of chitinase and chitosanase genes from T. harzianum could therefore result in a synergistic enhancement of anti-fungal activity of wheat.
Constitutive promoters have been used in maximum transgenic experiments in all the plants. In some experiments specially related to disease resistance the expressed protein is not required all the time or it is needed only in some specific parts of the plant. When it is expressed constitutively, there comes extensive cellular reprogramming of defence components in the parts of the plants which are not infected. This may give some times rise to plants which are resistant to diseases but weak in health and resulting productivity (Gurr and Rushton, 2005). For example in Arabidopsis NPR1 over expression using CaMV 35S promoter brought broad spectrum resistance with normal phenotype while in maize same was done using Ubiquitin promoter and it gave rise to diseased genotype without infection (Cao et al., 1998; Pieterse and Loon, 2004). Keeping in view these reasons and the fact that chitinase and chitosanase enzymes of the T. harzianum are highly antifungal it was decided to co-express HarChit and HarCho genes under constitutive as well as under inducible promoter. This way we could compare the impact of both the proteins on fungal diseases when they were present constitutively and induced under under pathogen attack. Vst-1 promoter from grape was wine as a stress inducible promoter that is also induced under disease in wheat (Leckband and Loerz, 1998). Ubiquitin promoter from Maize was used as constitutive promoter for the co-expression of HarChit and HarCho.
2.3 Performance of transgenics under constitutive promoter
A total of four lines having HarChit and HarCho, one line with HarCho and one transgenic control with gus gene under constitutive Ubi-1 promoter were evaluated against non transgenic control line Florida. Powdery mildew symptoms started to appear on the wheat segments 5-6 days after inoculation with E. graminis. Observations were taken on the number of developing colonies 9 dpi and colony size 21 dpi. The results showed a little or no decrease in the number of developing colonies per leaf segment on five lines and transgenic control line having Ubi-gus gene compared to the number of colonies per segment of leaf on non transgenic control (Figure 5). 9 dpi the difference in the size of the colonies for transgenic lines and controls was not visibly noticeable. 21 dpi the size of the fungal colonies was visibly different. On an average the fungal colonies on transgenic plants were 40-60% smaller than the colonies on transgenic and non transgenic controls. The colonies at the negative control lines were looking healthier with a lot of powdery mycelium while fungal colonies on transgenic leaves were mostly dead with very less powdery mycelium and the general appearance was brown and smaller colonies. Moreover a little more yellowishness was also observed in transgenic plants (Figure 6) compared to transgenics which may be an indication of accelerated Programmed cell death in the fight against fungus.
2.4 Performance of transgenics under Inducible promoter
The behaviour of the T1 progenies containing HarChit and HarCho genes under stress inducible gene was relatively different from transgenic progenies containing HarChit and HarCho under constitutive promoter. Out of the six T1 progenies (I.A-7-I.A-12) tested with E. graminis three showed almost the same number of colonies as control lines while the rest showed 8%-29% less colonies than the controls. Progeny I.A-8 had only HarCho but the resistance response of it against the development of E. graminis colonies was at par with other progenies having both the genes of interest. The size and health of the E. graminis colonies developing on the leaf disks of transgenics with the gene/genes of interest and control lines showed visibly smaller colonies with less powdery material while the leaf disks of the control lines showed visibly larger colonies with a lot of white powdery material. Out of the six lines tested, three lines (I.A-8, I.A-9 and I.A-10) showed the same size of colonies the size of colonies was observed on control lines. Three lines (I.A-7, I.A-11 and I.A-12) showed a reduction of 26, 75 and 36% in the colony size respectively compared to the control lines. The colonies on the controls were having more powdery material and were healthier than those found on the lines with over expressed HaChit and HarCho.
Various defence response genes have been used in wheat transformation for powdery mildew resistance. Oldach et al (2001) observed a reduction of 32%-40% in the number of developing E. graminis colonies on wheat transgenic line overexpressing Barley class-II chitinase and Ag-Afp protein from Aspergilos gigenteous. Our results indicate that chitinase and chitosanase proteins from T. harzianum worked in collaboration with endogenous plant defence response system and brought the transgenic to advantage as initially there was not a big difference in the number of developing colonies but the difference became visible afterwards in the health and size of colonies. If we talk about constitutive and inducible expression, it did not look to a major factor. Bieri et al., (2003) showed no reduction to less reduction in wheat susceptibility to E. graminis by high overexpression of RIP. Bieri et al., (2003) overexpressed barley seed antifungal proteins in wheat and checked the effect separately of alone chitinase, β-1,3-glucanse, RIP and Barnase as well as in combinations. RIP transgenics showed maximum reduction in powdery mildew susceptibility while chitinase and β-1,3-glucanse combination showed different levels of increase or decrease in susceptibility. A combination of three antifungal genes i.e. chitinase, RIP and β-1,3-glucanse produced by crossing did never show reduction of susceptibility better than the best parent. The little increase in the susceptibility of the lines I.A-8 and I.A-9 indicate that it is not necessary to quantitatively increase the anti-fungal proteins to increase resistance against powdery mildew disease rather a basal provision of anti-fungal proteins either produced constitutively or induced helps to increase disease resistance by working with endogenous plant defence system (Bieri et al., 2003). Sometimes this interaction does not bear results as is seen for lines I.A-8 and I.A-9 and seen by Bieri et al (2003).
3 Materials and Methods
3.1 Wheat transformation
For wheat transformation winter wheat genotype ‘Florida’ was selected. This genotype was maintained under standard conditions described by Leckband and Loerz, 1998, Becker et al., 1994 and Oldach et al., 2001. Tissue culture media, particle preparation, DNA precipitation and bombardment protocols was also followed from them. This genotype is highly responsive to tissue culture and readily transformable.
3.2 Expression vector construction
In total five expression cassettes were used in this study. Two of them containing HarChit and HarCho separately under the control of constitutive promoter Ubi1 from maize (Christensen et al., 1992) and Nopalin Synthase terminator from Agrobactrium tumefaciens were already cloned in the lab of Dirk Becker. Two of the expression vectors containing HarChit and HarCho separately were cloned stress cum disease inducible promoter from Vitis vinefera L. (Leckband and Loerz, 1998). In order to enhance the transcriptional activity of the Vst-1 promoter an enhancer fragment from the 35-S CaMV promoter was cloned 4 times at the 5′ end of Vst-1 promoter (Serazetdinova et al., 2005). Tnos was used as terminator. HarChit and HarCho were amplified from pUbiHarChit and pUbiHarCho plasmids using the primers given in table1.Vst1 and 4x Enhancer of 35-S promoter of CaMV were digested from pVst1EPG (Serazetdinova et al., 2005) plasmid, already present in the lab. All the five vectors are shown diagrammatically in Figure 7, Figure 8 and Figure 9.
 Figure 7 Vectors containing genes of interest under constitutive promoter. HarChit and HarCho under the Ubiquitin-1 promoter and nos terminator |
Figure 8 Vectors containing genes of interest under stress inducible promoter. HarChit and HarCho separately under the Vst-1 promoter and nos terminator. 4x Enhancer from CaMV, 35S Promoter is cloned on the upstream of Vst-1 to improve its expression level |
Figure 9 p35SAcS vector containing the pat selection marker gene under CaMV 35S promoter and 35S terminator. This vector was used in combination with both type of vectors conferring constitutive and inducible expression of HarChit and HarCho |
3.3 Molecular analysis of putative transgenics
The first step of transgenic analysis is to prove them at DNA level and then at RNA and lastly at protein level. Therefore all the BASTA selected plants were first checked by PCR and Southern analysis and then Northern Blot/rt PCR analysis was done. Lastly the activity of the overexpressed proteins was seen using pathological analysis with pathological fungi (Erysiphe graminis f.sp. tritici).
3.4 DNA isolati on and Southern Blot analysis
Genomic DNA was isolated by the protocols described by Palotta et al., 2000. Quality and quantity of isolated DNA was seen by using the spectrophotometer from Eppendorf, Germany and running DNA on agarose gel. DNA of each sample was divided into three portions; one was digested with endonuclease that cuts only once in the cassette, other was digested with endonuclease (s) cutting the expression cassette out of the plasmid, third portion was uncut. 25 µg from each portion was run on 0.8% agarose gel along with non transgenic wild type and plasmid DNA as negative and positive controls respectively. DNA was transferred onto Nylon membrane Hybobond NX (Ammersham Braunschweig Germany) by capillary method (Sambrook et al., 1989) and cross linked at 1250 J using Stratalinker TM 1800 UV Crosslinker (Stratagene, La Jolla, USA). The membrane transferred DNA was hybridized to the DIG labelled gene specific probes amplified by primer pairs given in table 2 and detected by CSPD substrate using the recommended protocols.
3.5 RNA Isolation and Northern Blot/rt-PCR
Freshly isolated total RNA was run on the 1 percent degeneration agarose gel in 1X MEN buffer. RNA was transferred to Hybond TM N+ Nylon membrane by capillary transfer using 10 X SSC solutions and fixed to membrane using the method mentioned above for Southern Blot analysis. For the detection hybridization positive single stranded gene specific DNA probes were radioactively labelled with P32 according to protocols given by manufacturers of DNA labelling kit (MBI Fermentas st. Leon-Rot, Germany). Detections were done on the X-ray films (Amersham, USA).
Northern blot analysis was done for transgenics where the genes (HarChit and HarCho) were constitutively expressed. The trangenics where the genes were under inducible promoter, the induction was done by rubbing the leaf surface with sea sand and total RNA was isolated eight hours afterwards (Leckband and Loerz, 1998 and Serazetdinova et al., 2005) cDNA was synthesized from the total RNA using 18-mer oligonucleotide (Oligo(dT)18) primer, dNTPs, RNAse inhibitor and the Moloney murine leukaemia virus reverse transcriptase (M-MuLV) as recommended by the reagents manufacturer (Fermentas Life Science, St. Leon Germany). Gene specific primers were used to amplify the transcripts of HarChit and HarCho out of cDNA from respective transgenic plants and lines.
3.6 Inoculation with Erysiphe graminis f sp. tritici
Inoculation studies with Erysiphe graminis f sp. tritici was performed on detached leaf segments using the method given below. All the pathological assays were revised at least thrice.
3.7 Detached leaves infections
In these experiments second leaf of two weeks old seedlings of T1 transgenic (Both over expression and knock down lines) and control lines (wild type Florida as well as transgenic line containing bar gene only) were cut into 3 cm long segments. These segments were cultured on an anti-senescence media containing 0.4% agar, 10 ppm (parts per million) benzamidazole and 1ppm silver nitrate. The protocols have already been used by Oldach et al., 2001 and Girgi et al., 2006. Inoculations were done in the settling tower by blowing the freshly harvested powdery fungal material mixed with equal volume of baby talcum powder onto the cultured leaf segments and at least one hour was given to settle the fungus down onto the plates. It was made sure that at least 300-400 conidia should be available per cm². Conidia were counted by putting a scale in the settling tower among plates at the time of inoculation and later on conidia were counted under microscope. Culture plates were covered with lids and sealed with sealing film and kept at 16℃ in the dark for overnight. After that these plates were transferred to infection chambers where a temperature of 18℃ and light to dark period ratio of 16:8 hours were maintained for the next 21 days. Fungus development was observed and number with size of developing colonies was taken.
Acknowledgements
We are highly thankful to DAAD, Germany and HEC, Pakistan for facilitating and funding the stay of a PhD student (1st Author) to do this research work. We are also thankful to Dr Kerstin Flath, of Plant Pathology and grassland Institute Kleinmachnow, Germany for providing fungal isolates and technical help to do the pathological analysis.
References
Anand A., Zhou T., Trick H.N., Gill B.S., Bockus W.W., and Muthukrishnan S., 2003, Greenhouse and field testing of transgenic wheat plants stably expressing genes for thaumatin-like protein, chitinase and glucanase against Fusarium graminearum, Journal of Experimental Botany, 54 (384): 1101-1111
http://dx.doi.org/10.1093/jxb/erg110 PMid:12598580
Balconi C., Lanzanova C., Conti E., Triulzi T., Forlani F., Cattaneo M., and Lupotto E., 2007, Fusarium head blight evaluation in wheat transgenic plants expressing the maize b-32 antifungal gene, European Journal of Plant Pathology, 117: 129-140
http://dx.doi.org/10.1007/s10658-006-9079-3
Becker D., Brettscheneider R., and Lörz H., 1994, Fertile transgenic wheat from microprojectile bombardment of scutellar tissue, The Plant Journal, 5: 299-307
http://dx.doi.org/10.1046/j.1365-313X.1994.05020299.x PMid:8148881
Becker D., Folk A., Knies P., Lörz H., and Wieser H., 2007, Silencing of alpha-Gliadins in Hexaploid Bread wheat, In: Georgr L.L.(eds.), Gluten Proteins, AACC International 2007
Bieri S., Potrykus I., and Futterer J., 2003, Effects of combined expression of antifungal barley seed proteins in transgenic wheat on powdery mildew infection, Molecular Breeding, 11: 37-48
http://dx.doi.org/10.1023/A:1022145915983
Blaisean P.L., and Lafay J.F., 1992, Primary structure of a chitinase-encoding gene (chi1) from the filamentous fungus Aphanocladium album: similarity to bacterial chitinases, Gene, 120: 243-248
http://dx.doi.org/10.1016/0378-1119(92)90099-B
Bliffeld M., Mundy J., Potrykus I., and Futterer J., 1999, Genetic engineering of wheat for increased resistance to powdery mildew disease, Theor. Appl. Genet., 98: 1079-1086
http://dx.doi.org/10.1007/s001220051170
Botha A.M., Nagel M.A.C., Van der Westhuizen A.J., and Botha F.C., 1998, Chitinase isoenzymes in near-isogenic wheat lines challenged with Russian wheat aphid, exogenous ethylene, and mechanical wounding, Bot Bull Acad Sin, 39: 99-106
Brettschneider R., Becker D., and Loerz H., 1997, Efficient transformation of scutellar tissue of immature maize embryos, Theor. Appl. Genet., 94: 737-748
http://dx.doi.org/10.1007/s001220050473
Cao H., Li X., and Dong X., 1998, Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance, Proc. Natl. Acad. Sci., USA, Plant Biology, 95: 6531–6536
Carsolio C., Benhamou N., Haran S., Cortés C., Gutiérrez A., Chet I., and Herrera-Estrella A., 1999, Role of the Trichoderma harzianum endochitinase Gene ech2 in Mycoparasitism, Applied and Environmental Microbiology, 65: 929-935 PMid:10049844 PMCid:91125
Chen W.P., Chen P.D., Liu D.J., Kynast R., Friebe B., Velazhahan R., Muthukrishnan S., and Gill B.S., 1999, Development of wheat scab symptoms is delayed in transgenic wheat plants that constitutively express a rice thaumatin-like protein gene, Theor. Appl. Genet., 99: 755-760
http://dx.doi.org/10.1007/s001220051294
Christensen A.H., Sharrock R.A., and Quail P.H., 1992, Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation, Plant Mol. Biol., 18: 675-689
http://dx.doi.org/10.1007/BF00020010 PMid:1313711
Girgi M., Breese W.A., Lörz H., and Oldach K.H., 2006, Rust and downy mildew resistance in pearl millet (Pennisetum glaucum) mediated by heterologous expression of the afp gene from Aspergillus giganteus, Transgenic Research, 15: 313-324
http://dx.doi.org/10.1007/s11248- 006-0001-8 PMid:16779647
Gurr S.J., and Rushton P.J., 2005, Engineering plants with increased disease resistance: How are we going to express it? Trends in Biotechnology, 23 (6): 283-290
http://dx.doi.org/10.1016/j.tibtech.2005.04.007 PMid:15922079
http://dx.doi.org/10.1016/j.tibtech.2005.04.009 PMid:15922080
Hammad H.M., Ahmad A., Laghari K.Q., Abbas F., Nasim W., Farhad W., and Malik A.H., 2011, Organic farming in wheat crop under arid condition of Pakistan, Pak J. Agri. Sci., 48(2): 97-102
Hendrix B., and Stewart J.M.D., 2002, Chitosanse may enhance anti-fungal defence response, AAES Research Series., 507: 2154-217
Khan S., and Khan J., 2010, Drought tolerant wheat cultivar (raj) for rainfed areas of KPK, Pakistan, Pak J. Agri. Sci., 47(4): 355-359
Leckband G., and Loerz H., 1998, Transformation and expression of a stilbene synthase gene of Vitis vinifera L. in barley and wheat for increased fungal resistance, Theor. Appl. Genet., 96 : 1004-1012
http://dx.doi.org/10.1007/s001220050832
Lee S., Choi H., Suh S., Doo I.S., Oh K.Y., Choi E.J., Schroeder Taylor A.T., Low P.S., and Lee Y., 1999, Oligogalacturonic acid and chitosan reduce stomatal aperture by inducing the evolution of reactive oxygen species from guard cells of tomato and Commelina communis, Plant Physiology, 121: 147-152 http://dx.doi.org/10.1104/pp.121.1.147 PMid:10482669 PMCid:59362
Lorito M., Peterbauer C., Hayes C.K., and Harman G.E., 1994, Synergistic interaction between fungal cell wall degrading enzymes and different antifungal compounds enhances inhibition of spore germination, Microbiology, 140 (3): 623-629http://dx.doi.org/10.1099/00221287-140-3-623
PMid:8012584
Lorito M., Farkas V., Rebuffat S., Bodo B., and Kubicek C.P., 1996, Cell wall synthesis is a major target of mycoparasitic antagonism by Trichoderma harzianum, J. Bacteriol., 178(21): 6382-6385 PMid:8892847 PMCid:178518
Mackintosh C.A., Lewis J., Radmer L.E., Shin S., Heinen S.J., Smith L.A., Wyckoff M.N., Dill-Macky R., Evans C.K., Kravchenko S., Baldridge G.D., Zeyen R.J., and Muehlbauer G.J., 2007, Overexpression of defence response genes in transgenic wheat enhances resistance to Fusarium head blight, Plant Cell Reports., 26: 479-488
http://dx.doi.org/10.1007/s00299-006-0265-8 PMid:17103001 PMCid:1824786
Makandar R., Essig J.S., Schapaugh M.A., Trick H.N., and Shah J., 2006, Genetically engineered resistance to Fusarium head blight in wheat by expression of Arabidopsis NPR1, Mol Plant Microbe Interact., 19: 123-129 http://dx.doi.org/10.1094/MPMI-19-0123 PMid:16529374
Muehlbauer G.J., and Bushnell W.R., 2003, Transgenic approaches to resistance, In: Leonard K.J., and Bushnell W.R. (eds), Fusarium head blight of wheat and barley, American Phytopathological Society Press, St. Paul, MN
Naz A.A., 2007, Comparative mapping of genes for plant disease resistance in wheat advanced ‘backcross populations’ by means of DNA markers, PhD thesis, Faculty of Agriculture, University of Bonn, Germany
Oldach K.H., Becker D., and Loerz H., 2001, Heterologous expression of genes mediating enhanced fungal resistance in transgenic wheat, Molecular Plant–Microbe Interactions, 14: 832-838
http://dx.doi.org/10.1094/MPMI.2001.14.7.832 PMid:11437256
Palotta M.A., Graham R.D., Langridge P., Sparrow D.H.B., and Barker S.J., 2000, RFLP mapping of manganese efficiency in barley, Theor Appl Genet, 101: 1100-1108
http://dx.doi.org/10.1007/s001220051585
Papavizas G.G., 1985, Trichoderma and Gliocladium: Biology, ecology and potential for biocontrol, Ann Rev Phytopathol, 23: 23-54
http://dx.doi.org/10.1146/annurev.py.23.090185.000323
Pellegrineschi A., Noguera L.M., Skovmand B., Brito R.M., Velazquez L., Salgado M.M., Hernandez R., Warburton M., and Hoisington D., 2002, Identification of highly transformable wheat genotypes for mass production of fertile transgenic plants, Genome, 45: 421-430
http://dx.doi.org/10.1139/g01-154 PMid:11962639
Pieterse C.M.J., and Loon L.C.V., 2004, NPR1: the spider in the web of induced resistance signalling pathways, Current Opinion in Plant Biology, 7: 456-464
http://dx.doi.org/10.1016/j.pbi.2004.05.006 PMid:15231270
Sambrook J., Fritsch E.F., and Maniatis T., eds., 1989, Molecular cloning: a laboratory manual, 2nd edn, Cold Spring Habour Laboratory, Cold Spring Harbor, New York
Serazetdinova L., Oldach K.H., and Loerz H., 2005, Expression of transgenic stilbene synthases in wheat causes the accumulation of unknown stilbene derivatives with antifungal activity, J. Plant Physiol., 65: 985-1002 http://dx.doi.org/10.1016/j.jplph.2004.11.005 PMid:16173460
Shin S., Mackintosh C.A., Lewis J., Heinen S.J., Radmer L., Dill-Macky R., Baldridge G.D., Zeyen R.J., and Muehlbauer G.J., 2008, Transgenic wheat expressing a barley class II chitinase gene has enhanced resistance against Fusarium graminearum, J. Exp. Bot., 59: 2371-2378
http://dx.doi.org/10.1093/jxb/ern103 PMid:18467324 PMCid:2423652
Singh A., Isuas Kirubakavan S., and Sakthivel N., 2007, Heterologous expression of new antifungal chitinase from wheat, Protein Expression and Purification, 56: 100-109
http://dx.doi.org/10.1016/j.pep.2007.06.013 PMid:17697785
Sivamani E., Bahieldin A., Wraith J.M., Al-Niemi T., Dyer W.E., Ho T.D.H., and Qu R., 2000, Improved biomass productivity and water use efficiency under water deficit conditions in transgenic wheat constitutively expressing the barley HVA1 gene, Plant Science, 155:1-9
http://dx.doi.org/10.1016/S0168-9452(99)00247-2
Steyaert J.M., Stewart A., Jaspers M.V., Carpenter M., and Ridgway H.J., 2004, Co-expression of two genes, a chitinase (chit42) and proteinase (prb1), implicated in mycoparasitism by Trichoderma hamatum, Mycologia, 96: 1245-1252
http://dx.doi.org/10.2307/3762141 PMid:21148948
Vander P., Vrum K.M., Domard A., Eddine E., Gueddari N., and Moerschbacher B.M., 1998, Comparison of the ability of partially N-acetylated chitosans and chitooligosaccharides to elicit resistance reactions in wheat leaves, Plant Physiol., 118(4): 1353-1359
http://dx.doi.org/10.1104/pp.118.4.1353 PMid:9847109 PMCid:34751
Yanai K., Takaya N., Kojima H., Horiuchi A., and Ohta M.T., 1992, Purification of two chitinases from Rhizopus oligosporus and isolation and sequencing of the encoding genes, J. Bacteriol., 174: 7398-7406 PMid:1429462 PMCid:207436
. PDF(764KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Iqrar Ahmad Rana
. Horst Loerz
. Wilhelm Schaefer
. Dirk Becker
Related articles
. Inducible and constitutive promoter
. Chitinase
. Chitosanase
. Vst -1
. Ubiquitin-1
. Wheat
. Transformation
Tools
. Email to a friend
. Post a comment


