Author
Author  Correspondence author
Correspondence author
Molecular Plant Breeding, 2019, Vol. 10, No. 6 doi: 10.5376/mpb.2019.10.0006
Received: 11 Mar., 2019 Accepted: 25 Mar., 2019 Published: 29 Mar., 2019
Ren Y., Lu J.H., Huang S.H., Zhang Z.Q., Kong X., 2019, QTLs location of calyx-related traits in tetraploid Dendrobium, Molecular Plant Breeding, 10(6): 43-49 (doi: 10.5376/mpb.2019.10.0006)
In order to identify QTL locus of calyx-related traits in Dendrobium, 190 F1 individuals derived from the crossbreed of tetraploid Dendrobium cultivars D. Mangosteen x D. Burana Pink No.2 were used as materials. Through three years observation of four characteristics of calyx (length of dorsal sepal, width of dorsal sepal, length of lateral sepal, width of lateral sepal), the QTL was located on the two genetic maps of parents which had been already constructed. The result showed that the four characteristics were quite different among the individuals of F1 generation, and the observed values of all the characteristics were all positive distribution, which was suitable for QTL analysis. Nine QTLs were detected on the maternal ‘D. Mangosteen’ genetic linkage map, including 1 QTL for dorsal sepal length, 3 QTLs for dorsal sepal width, 2 QTLs for lateral length and 3 QTLs for lateral width, with the genetic contribution rate ranging from 11.9% to 16.8%. Also, two tight linkage markers were acquired (M10E3-146, M8E8-284). A total of 6 QTLs were detected on the paternal ‘D. Burana Pink No.2’ linkage map, including 2 QTLs for dorsal sepal length, 1 QTL for dorsal sepal width, 2 QTLs for lateral length and 1 QTL for lateral width, with the genetic contribution rate ranging from 11.8% to 17.3%. This result of study could provide reference for Dendrobium molecular marker assisted breeding and precise location of correlation gene.
Background
Dendrobium is a kind of perennial herb belongs to the genus Dendrobium of the Orchidaceae, which is an ornamental with bright market prospect. Among tropical orchid, it is listed as “the top four ornamental foreign orchid” with Cattleya, Phalaenopsis, and V anda for their high ornamental value (Zeng and Hu, 2004). At present, the breeding of dendrobium is in traditional way. It costs long time and being inefficient. The construction of high-density genetic map and QTL of related traits could improve breeding efficiency, thus shortening breeding time (He et al., 2017). Recent years, the research for Dendrobium focused on panting (Sun et al., 2012; Jiang, 2014), tissue culture (Zhao et al., 2012; Jiang, 2014; Wang et al., 2017) and genetic diversity and relationships (Bai et al., 2007; Ren et al., 2013). Because of ploidy’ complexity, which is presented as diploid, triploid, tetraploid and hexaploid, most of the species are diploid (2n=2x=38) (Jones et al., 1998), most of the cultivars are tetraploid (2n=4x=76) (Liao et al., 2012), so the process of dendrobium genetic map construction research is slow.
At present, the research of Dendrobium genetic map consruction mainly focused on diploid, Xue et al., (2010) used 90 F1 individuals of diploid ‘Dendrobium candidum’ × ‘Dendrobium hercoglossum’as mapping population, constructing a parental genetic map respectively by using RAPD and SRAP mark. Lu et al., (2012a) used 90 F1 individuals of diploid ‘Dendrobium moniliforme’ × ‘Dendrobium candidum’ as construction material, constructing a parental genetic map respectively by using EST-SSR, SRAP, ISSR and RAPD mark. In the same year, Lu et al. (2012b) used 90 F1 individuals of diploid ‘Dendrobium candidum’ × ‘Dendrobium aduncum’ as construction material, constructing an integrated genetic map by using EST-SSR and SRAP mark. Feng et al., (2013) used 90 F1 individuals of diploid ‘Dendrobium nobile’ × ‘Dendrobium moniliforme’ as construction material, constructing a parental genetic map respectively by using RAPD and ISSR mark. Huang (2007) used 90 F1 Dendrobium nobile hybrid individuals of Den. Lucky Gal ‘Emito’ × Den. Fantsy ‘Crown’ as construction material, constructing a Dendrobium nobile genetic linkage map by using RADP mark. However, no studies have been reported on QTL location of related traits in Dendrobium. By using SRAP, RSAP, SSR, ISSR and tetraploid Dendrobium (2n=4x=76) as mapping population, the research constructed a parental genetic map respectively. The research aimed to conduct positioning for related traits of calyx on the basis of Dendrobium Phalaenopsis complete parental genetic map, which provided references for gene cloning of important traits in Dendrobium, accurate positioning and molecular marker assisted breeding.
1 Results and Analysis
1.1 Analysis of main characters of F1 population
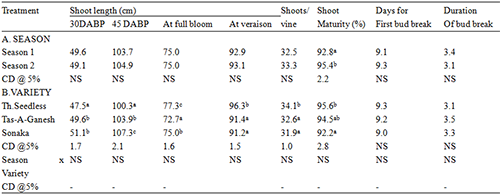
Statistics was conducted for the related traits of 190 individuals’ sepal in F1 population. It is found that there are obvious differences in the length of dorsal sepal, the width of dorsal sepal, the length of lateral sepal and the width of lateral sepal. Continuous variation of statistic was presented as characters of quality and traits. The absolute values of skewness and kurtosis are all less than 1. All traits conform to normal distribution (Figure 1; Table 1).
|
Figure 1 The frequency distribution of 4 traits in F1 population of Dendrobium |
|
Table 1 Parameters of 4 traits in F 1 population |
1.2 QTL analysis of calyx traits
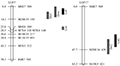
Based on the constructed genetic map, we combined phenotypic data, used MapQTL 6 software to conduct QTL location analysis. Choosing LOD ≥2.5 as critical value of QTL, nine QTLs with four traits of dorsal sepal length, dorsal sepal width, lateral sepal length and lateral sepal width were detected in the linkage group of female parents, which distributed in five linkage groups. Six QTL were detected in the male parent linkage group, which were distributed in 2 linkage groups (Table 2; Figure 2; Table 3; Figure 3).
|
Table 2 The distribution and effect of QTL locus in D. Mangosteen |
|
Figure 2 QTL position of female parent D. Mangosteen on linkage group |
|
Table 3 The distribution and effect of QTL locus in D. Burana Pink No.2 |
|
Figure 3 QTL position of male parent D. burana Pink No.2 on linkage group |
1.2.1 QTL characteristics of maternal traits
Length of dorsal sepal: 1 QTL was detected in linkage group of LGM16, and the location on the linkage group was 86.8 cM, whose genetic contribution rate was 16.8%. The distance between the location and the closest marker was 3 cM (Table 2).
Width of dorsal sepal: 3 QTL were detected in linkage groups of LGM 10, LGM13 and LGM21. The locations on the linkage groups were 32.8 cM, 27 cM and 41.6 cM, whose genetic contribution rates were 12.7%, 12.4% and 14.6%, respectively. The distances between the location and the closest marker were 2.0 cM, 2.0 cM and 7.0 cM (Table 2).
Length of lateral sepal: 2 QTL were detected in linkage group of LGM16, the locations on the linkage group were 26.5 cM and 85.8 cM, whose genetic contribution rates were 12.6% and 14.8%. The distances between the locations and the closest marker were 5.2 cM and 2.0 cM (Table 2).
Width of lateral sepal: 3 QTL were detected, of which one was on LGM15 linkage group and two were on LGM16 linkage group, and the locations on the linkage group were 31.9 cM, 60.0 cM and 81.5 cM, whose genetic contribution rates were 14.8%、12.0% and 11.9%, respectively. The distance between Lsw1 in LGM15 linkage group and the closest marker M10E3-146 and Lsw2 in LGM16 linkage group and the closest marker M8E8-284 was 0. Which indicated that the two markers and locations were closely linked (Table 2).
1.2.2 QTL characteristics of paternal traits
Length of dorsal sepal: 2 QTL were detected in linkage groups of LGF5 and LGF17, the locations on the linkage group were 10.0 cM and 42.0 cM, whose genetic contribution rates were 12.5% and 17.3%. The distances between the location and the closest marker were 4.1 cM and 5.7 cM (Table 3).
Width of dorsal sepal: 1 QTL was detected in linkage group of LGF5; the location on the linkage group was 4.0 cM, whose genetic contribution rate was 14.5%. The distance between the location and the closest marker was 4.0 cM (Table 3).
Length of lateral sepal: 2 QTL were detected in linkage group of LGF5 and LGF17, the locations on the linkage group were 5.0 cM and 39.0 cM, whose genetic contribution rate were 12.8% and 11.9%. The distances between the location and the closest marker were 5.0 cM and 8.7 cM.
Width of lateral sepal: 1 QTL was detected in linkage group of LGF5, the locations on the linkage group was 24.1 cM, whose genetic contribution rate was 11.8%. The distance between the location and the closest marker was 1.7 cM, which indicated that the marker and the location were closely linked (Table 3).
2 Discussion
Dendrobium is a kind of flower with high ornamental value, and its flower is the most important parts for ornamental and it is the significant goal of its breeding. At present, there is no report on the location of QTL in Dendrobium orchids. In the research of tetraploid plants’ genetic mapping and QTL location, the potato was mostly reported. Bradshaw et al. (2008) found variation of QTL’s number, location and distribution of traits in paternal map in the locating studies of yield, maturity and quality traits of tetraploid potato. Judged from that, the influence of target traits on offspring is also different. Cui et al. (2016) also found similar phenomenon in QTL mapping analysis of three important traits of anthocyanin content, tuber yield per plant and marketable tuber percentage of tetraploid colored potato. It was also suggested that the QTL alleles of the offspring could be analyzed to judge which part of parent had the advantage by the genetic information of the parents. This study had similar results, QTL of 4 characters related to Dendrobium calyx was located on the basis of constructing the parents' genetic map by using F1 group respectively. It was found that 9 QTL locations were detected on the female parent maps of 4 maternal traits, distributing in 5 linkage groups and 6 QTL locations were detected on the male parent map, distributing in 2 linkage groups. The results showed that the number, location and distribution of QTLs detected by these four traits were different in the parent map of Dendrobium.
The results also indicated there is a phenomenon called common region between linkage groups of four traits. For example, Lsw3 location of lateral sepal width and Dsl location of dorsal sepal length and Lsl2 location of lateral sepal length detected in the maternal map were in continuous extents; with Dsl location of dorsal sepal length and Lsl2 location of lateral sepal length were located in the same region of R6R10-1121-M10E5-96 on 16 linkage groups (Table 2); However, two locations of dorsal sepal length and two locations of lateral sepal length detected on male parent map were all located in the same segment (Table 3). It is indicated that these two traits might have a close relationship. There was similar phenomenon in potatoes (Schafer et al., 1998; Cui etal., 2016) and peanuts (Chen et al., 2015).
It is also found that QTL clustering is common in different linkage groups. For example, three traits of the dorsal sepal length, the lateral sepal length and the lateral sepal width, appeared in clusters in the 16th linkage group on the maternal map (Figure 2). Four traits of the dorsal sepal length, the dorsal sepal width, the lateral sepal length and the lateral sepal width, clustered on the 5th linkage group on the paternal map (Figure 3). Multi-trait QTL clusters also exist in other crops (Portis et al., 2014; Chen et al., 2015; Ge et al., 2015), it may be the phenotypically related traits were regulated by one or a few polygenic genes and co-segregated genetically. This can effectively increase gene utilization and reduce gene loss caused by recombination (Liu et al., 2013; 2016). Furthermore, the distance between Lsw1, Lsw2 location of lateral sepal width and adjacent markers M10E3-146 and M8E8-284 was 0, these 2 loci could be thought to be closely linked to molecular markers. This provided references for fine mapping and cloning of QTL gene and molecular marker assisted breeding.
3 Materials and Methods
3.1 Materials
190 F1 individuals of tetraploid D. Mangosteen × D. Burana Pink No.2 were used as experimental material. The material was planted in experiment base of Tropical Crops Genetic Resources Institute, Chinese Academy of Tropical Agricultural Sciences. Each individual was panted in 12 cm×18 cm plastic basin, respectively.
3.2 Observation of agronomic traits
Choosing 190 F1 individuals between 2014 and 2016, conducting three-year observation and records on the basis of Descriptors and Data Quality Control for Dendrobium(Dendrobium. Sw) (Yin et al., 2008).
3.3 Data statistics and analysis methods
Statistical analysis was made on the observed values of four characters of the dorsal sepal length, the dorsal sepal width, the lateral sepal length and the lateral sepal width by SAS software. QTL mapping was carried out by using MapQTL 6.0 software on the constructed Genetic linkage map of the parents of tetraploid Dendrobium, according to the measurement results of four Calyx traits. Interval mapping was used to analyze genotypes and traits. Software automatically searched for QTL associated with molecular markers when LOD is ≥ 2.5, drawing QTL map by MapChart 2.2 software (Voorrips, 2006). The nomenclature of the QTL location was represented by “the abbreviation of traits +QTL number”.
Authors’ contributions
HSH, ZZQ, KX and LJH were the designers of this experiment and the executors of this experiment and research; LJH was responsible for data analysis, results analysis of the experiment and writing of the first draft of the paper; RY is responsible for the conceiving and he was responsible person of the project, he guided experimental design, data analysis, thesis writing and revision. All authors read and approved the final manuscript.
Acknowledgments
The research was jointly financed by Central Public-interest Scientific Institution Basal Research Fund for Chinese Academy of Tropical Agricultural Sciences (1630032017021; 1630032017024).
Bai Y., Bao Y.H., Wang Q.W., Jiang L.L., and Yan Y.N., 2007, Analysis of the phyloge netic relationship of Dendrobium in China by AFLP technique, Yuanyi Xuebao (Acta Horticulturae Sinica), 34(6): 1569-1574
Bradshaw J.E., Hackett C.A., Pande B., Waugh R., and Bryan G.J., 2008, QTL mapping of yield, agronomic and quality traits in tetraploid potato (Solanum tuberosum subsp. tuberosum), Theor. Appl. Genet., 116(2): 193-211
https://doi.org/10.1007/s00122-007-0659-1
PMid:17938877
Cheng L.Q., Tang M., Ren X.P., Huang L., Chen W.G., Li Z.D., Zhou X.J., Chen Y.N., Liao B.S., and Jiang H.F., 2015, Construction of genetic map and QTL analysis for mainstem height and total branch number in peanut (Arachis hypogaea L. ), Zuowu Xuebao (Acta Agronomica Sinica ), 41(6): 979-987
https://doi.org/10.3724/SP.J.1006.2015.00979
Cui K.S., Yu X.X., Yu Z., Jiang C., and Shi Y., 2016, QTL location of anthocyanin content and yield in tetraploid pigmented potato, Caoye Xuebao (Acta Prataculturae Sinica), 25(5): 116-124
Feng S., Zhao H., Lu J., Liu J., Shen B., and Wang H., 2013, Preliminary genetic linkage maps of Chinese herb Dendrobium nobile and D. moniliforme, Journal of Genetics, 92 (2):205-212
https://doi.org/10.1007/s12041-013-0246-y
PMid:23970076
Ge H.Y., Liu Y., and Chen H.Y., 2015, QTL analysis of fruit-associated traits in eggplant, Yuanyi Xuebao (Acta Horticulturae Sinica), 42(11): 2197-2205
He D., Lei Y.K., Wang Z., Liu Y.P., Zhang M., and He S.L., 2017, Research progress on genetic linkage map construction and QTL mapping of flowers, Fenzi Zhiwu Yuzhong (Molecular Plant Breeding), 15(3): 994-1002
Huang S.L., 2007, Identification chromosome ploidy of dendrobium and construction of RAPD molecular genetic maps, Thesis for M.S., Huazhong Agricultural University, Supervisor: Bao M.Z., pp.26-38
Jiang H.T., 2014, Research on key techniques of seedling rapid propagation and cultivation of Demdrobium officinale, Thesis for M.S., Central South University of Forestry and Technology, Supervisor: Zhang L., He J.X., pp.10-18, 19-30
Jones W.E., Kuehnle A.R., and Arumuganathan K., 1998, Nuclear DNA content of 26 orchid (Orchidaceae) genera with emphasis on Dendrobium, Annals of Botany, 82(2): 189-194
https://doi.org/10.1006/anbo.1998.0664
Liao D.L., Xie L., Zeng R.Z., Li Y.H., Yi M.S., and Zhang Z.S., 2012, Relationship between chromosome ploidy and morphology characters in Dendrobium, Xibei Zhiwu Xuebao (Acta Botanica Boreali-Occidentalia Sinica), 32(10): 2023-2029
Liu D.X., Zhang J., Liu X.Y., Wang W.W., Liu D.J., Teng Z.H., Fang X.M., Tan Z.Y., Tang S.Y., Yang J.H., Zhong J.W., and Zhang Z.S., 2016, Fine mapping and RNA-Seq unravels candidate genes for a major QTL controlling multiple fiber quality traits at the T1 region in upland cotton, BMC Genomics, 17: 295
https://doi.org/10.1186/s12864-016-2605-6
PMid:27094760 PMCid:PMC4837631
Liu T.M, Yu T., and Xing Y.Z., 2013, Identification and validation of a yield-enhancing QTL cluster in rice (Oryza sativa L.), Euphytica, 192(1): 145-153
https://doi.org/10.1007/s10681-012-0796-8
https://doi.org/10.1007/s10681-013-0929-8
Lu J.J., Zhao H.Y., Suo N.N., Wang S., Shen B., Wang H.Z., and Liu J.J., 2012a, Genetic linkage maps of Dendrobium moniliforme and D. officinale based on EST-SSR, SRAP, ISSR and RAPD markers, Scientia Horticulturae, 137(1): 1-10
https://doi.org/10.1016/j.scienta.2011.12.027
Lu J.J., Wang S., Zhao H.Y., Liu J.J., and Wang H.Z., 2012b,Genetic linkage map of EST-SSR and SRAP markers in the endangered Chinese endemic herb Dendrobium (Orchidaceae), Genetics and Molecular Research, 11(4): 4654-4667
https://doi.org/10.4238/2012.December.21.1
PMid:23315811
Portis E., Barchi L., Toppino L., Lanteri S., Acciarri N., Felicioni N., Fusari F., Barbierato V., Cericola F., Vale G., and Rotino G.L., 2014, QTL mapping in eggplant reveals clusters of yield-related loci and orthology with the tomato genome, PLoS One, 9(2): e89499
https://doi.org/10.1371/journal.pone.0089499
PMid:24586828 PMCid:PMC3931786
Ren Y., Yang G.S., Lu S.J., Xu S.S., Huang S.H., and Yin J.M., 2013, Analysis of genetic diversity of dendrobium by RSAP marker, Agricultural Science & Technology, 14(12): 1710-1713, 1722
Schafer P.R., Ritter E., Concilio L., Hesselbach J., Lovatti L., Walkemeier B., Thelen H., Salamini F., and Gebhardt C., 1998, Analysis of quantitative trait loci (QTLs) and quantitative trait alleles (QTAs) for potato tuber yield and starch content, Theor. Appl. Genet., 97(5-6): 834-846
https://doi.org/10.1007/s001220050963
Sun C.X., Feng M.L., Chen H., and Chen W.J., 2012, Effects of different coconut husk chip treatments on the growth of Anthurium and Dendrobium sp., Redai Nongye Kexue (Chinese Journal of Tropical Agriculture), 32(1): 1-4
Voorrips R.E., 2006, Map Chart: Software for the graphical presentation of linkage maps and QTLs, Journal of Heredity, 93(1): 77-78
https://doi.org/10.1093/jhered/93.1.77
Wang Z.Q., Meng J.L., and Cui Y.Y., 2017, Tissue culture of soot apices for induction of PLBs of Dendrobium nobile ‘Huoniao’, Zhejiang Linye Keji (Journal of Zhejiang for Science & Technology), 37(2): 48-54
Xue D.W., Feng S.G., Zhao H.Y., Jiang H., Shen B., Shi N.N., Lu J.J., Liu J.J., and Wang H.Z., 2010, The linkage maps of Dendrobium species based on RAPD and SRAP markers, J. Genet. Genomics, 37(3): 197-204
https://doi.org/10.1016/S1673-8527(09)60038-2
Yin J.M, Ren Y., Yang G. S. Descriptors and data quality control for dendrobium (Dendrobium.Sw), 2008, China Agriculture Press, pp.52-80
Zhao X.B., Wu W.J., Pang L., and Cai G.X., 2012, Study on key technology of rapid culture and propagation for Dendrobium candidum as rare and endangered medicinal materials, Hunan Zhongyiyao Daxue Xuebao (Journal of Traditional Chinese Medicine University of Hunan), 32(3): 27-30
. PDF(424KB)
. FPDF
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Yu Ren
. Jinghua Lu
. Shaohua Huang
. Zhiqun Zhang
. Xia Kong
Related articles
. Tetraploid Dendrobium
. Calyx-related traits
. QTL location
Tools
. Email to a friend
. Post a comment