Research Report
Genome-wide Identification and Bio-informatics Analysis of Superoxide Dismutase Gene Family in Zea mays 
2 Agricultural College, Northeast Agricultural University, Harbin, 150030, China
3 Agriculture and Forestry Science and Technology College, Hebei North University, Zhangjiakou, 075000, China
 Author
Author  Correspondence author
Correspondence author
Molecular Plant Breeding, 2019, Vol. 10, No. 8 doi: 10.5376/mpb.2019.10.0008
Received: 21 Mar., 2019 Accepted: 22 May, 2019 Published: 15 Jul., 2019
Guo D., Du M., Xu H.L., Zhou B.Y., Ma W., Ding Z.S., Li C.F., and Zhao M., 2019, Genome-wide identification and bio-informatics analysis of superoxide dismutase gene family in Zea mays, Molecular Plant Breeding, 10(8): 59-67 (doi: 10.5376/mpb.2019.10.0008)
Superoxide dismutase (SOD) is an important antioxidant enzyme and widely exists in organisms. The whole genome scanning and bioinformatics analysis of millet superoxide dismutase (SOD) gene family were carried out by using bioinformatics method on the search platform of maize gene database. The results showed that the SOD gene family of maize contained 15 genes, which were unevenly distributed on 9 chromosomes. The amino acid sequence length of the protein ranged from 86 aa to 386 aa. Fifteen SOD proteins in maize contained 6 Cu-SOD motifs and 9 Mn-SOD motifs. Evolutionary tree analysis showed that SOD protein of maize was related to SOD protein sequence of millet and sorghum. The results of this study can provide some references for further understanding the SOD gene family and the antioxidant mechanism of maize.
Maize is a kind of warm temperate crop, which is originated from tropical areas and is an important food crop and cash crop. However, the growth and yield of maize are constantly affected by abiotic and biological stresses, such as pathogenic bacteria infection, drought and flooding, metal ions, chemical agents, etc. (Chen et al., 2018). It has been found that these environmental factors can lead to the production and increase of reactive oxygen species (ROS) in plant cells, while excessive ROS can lead to damage to plant cells, and may even lead to irreparable metabolic dysfunction and cell death (Lee et al., 2007; Karuppanapandian et al., 2011). Therefore, in order to cope with ROS excess, plants have formed complex non-enzymatic and enzymatic antioxidant protection systems to eliminate ROS in the long-term evolution process. Among these enzymes, superoxide dismutase (SOD) is the first defense line of plant defense system, a metal enzyme widely existing in organisms, and a key enzyme for scavenging oxygen free radicals in plants. Its activity can affect plant growth and development (Rizhsky et al., 2003), structural and functional stability of cell membrane (Allen et al., 1997) and plant stress resistance (Guo et al., 2005). Close correlation.
As early as 1969, Fridovich and MeCord discovered that a blood-copper protein can scavenge free radicals and is named superoxide dismutase (SOD) (Mccord and Fridovich, 1969). According to the difference of metal ions in the active center of the enzyme, it can be divided into Cu/Zn-SOD, Fe-SOD, Mn-SOD and Ni-SOD (Zelko et al., 2002). Different superoxide dismutases are distributed in different sub-Asianes. In the cell structure, Cu/Zn-SOD is present in chloroplast (Palma et al., 1986), cytosol (Bannister et al., 1987) and mitochondria (Baum and Scandalios, 1981); Mn-SOD is present in mitochondria and peroxisomes (Duke and Salin, 1985; Sandalio and Del, 1987); Fe- SOD is mainly found in chloroplasts and can also be detected in mitochondria and peroxisomes (Droillard and Paulin, 1990). Ni-SOD is mainly present in the cytoplasm (Youn et al., 1996). Among them, Cu/Zn-SOD is especially important in the enzyme system of active oxygen scavenging, and it is closely related to various drought resistance, cold resistance, salt and alkali resistance of plants (Feng et al., 2005; Zhang et al., 2005; Song et al., 2006). Transgenic tobacco research found that overexpression of Cu/Zn-SOD can reduce the damage of water stress on plants (Faize et al., 2011). Fe-SOD and Mn-SOD are mainly related to plant stress resistance. In the physiological and biochemical studies of tobacco and alfalfa plants, the cold resistance and antioxidant capacity of the plants were significantly enhanced (Wim et al., 1996; Bryan et al., 2000).
Although the SOD study has a history of nearly 100 years (Faize et al., 2011), the maize SOD gene family has not been reported. In this paper, we carried out genome-wide mining and analysis of maize SOD gene family, and analyzed the basic information, conserved domain structure, evolutionary relationship and chromosomal location of maize SOD gene family to provide information for maize SOD gene cloning and to study corn antioxidant activity. The mechanism provides a certain reference.
1 Results and Analysis
1.1 Identification and gene mapping of SOD gene family in Maize
The hidden Markov model of SOD family (Pfam number: PF00080, PF00081, PF02777) was downloaded from the Pfam database, and the Pfam number of SOD protein was searched for similar sequences. A total of 15 SOD protein sequences were obtained. The protein sequence domains were detected by SMART and CDD, and the deletion and redundancy were checked. Finally, 15 SOD gene sequences were obtained. According to their chromosomal location, they were named ZmSOD1 to ZmSOD15 (Table 1). Analysis of 15 SOD gene sequences in maize showed that there were significant differences among different genes: the length of amino acids encoding SOD protein ranged from 86aa (ZmSOD9) to 386aa (ZmSOD13), the open reading frame ranged from 1279 (ZmSOD9) to 13650 (ZmSOD11), the molecular weight ranged from 9.81 (ZmSOD9) to 42.86 (ZmSOD13), and the isoelectronic range ranged from 5.42 (ZmSOD5) to 8.98 (ZmSOD7).
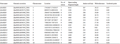 Table 1 Information of SOD-domain gene family in maize |
From Table 1 and Figure 1, it was found that SOD genes in maize were unevenly distributed on chromosomes 8, of which chromosome 1 had up to 4 genes, chromosome 6 and chromosome 9 had 3 genes, and other chromosomes had 1 to chromosome respectively. The results of gene replication analysis showed that family genes were distributed at both ends of chromosome and few near centromere, so SOD genes were far apart on chromosome. The amplification of gene family could not be achieved by tandem repetition, but the distribution of genes on chromosome was related to the growth and development of maize.
 Figure 1 Chromosome mapping of SOD genes in maize |
1.2 Domain analysis of SOD protein in Maize
The domain of 15 maize SOD proteins was analyzed using ProSite software (Figure 2). The results showed that the maize SOD protein mainly contains two domains: one is the Cu/Mn-SOD domain; the other is composed of the N-terminal C domain and the C-terminal Fe/Mn-SOD domain. ZmSOD1, ZmSOD2, ZmSOD3, ZmSOD6, ZmSOD10, and ZmSOD14 proteins contain Cu/Mn-SOD domains, and ZmSOD5, ZmSOD7, ZmSOD8, ZmSOD11, ZmSOD12, ZmSOD13, and ZmSOD15 proteins contain Fe/Mn-SOD and Cu/Mn-SOD domains. There are two special proteins containing only one Fe/Mn-SOD and Cu/Mn-SOD domains, namely: ZmSOD4 and ZmSOD14.
 Figure 2 The domain of SOD protein in Maize Note Brown represents the Cu/Mn domain; Blue represents the N terminal Fe/Mn domain; Green represents the C terminal Fe/Mn domain |
The conserved motifs of 15 SOD protein sequences in maize were predicted by MEME online website. As shown in Figure 3, the number and type of conserved motifs contained in each protein sequence were different. The difference in the distribution of conserved motifs might reveal the different functions of each gene. It can be seen that ZmSOD1, ZmSOD2, ZmSOD3, ZmSOD10 and ZmSOD12 all contain motif 2; motif 1, motif 3, motif 4 and motif 5 are distributed on other protein sequences respectively, which may play an important role in plant growth and development or stress. It is learned from table 2 that motif 2 has a long sequence length of 74 and can be used to analyze the 3D structure of SOD protein.
 Figure 3 Analysis of SOD protein motif in Maize |
 Table 2 Conserved motif of SOD protein in Maize |
The SWS-MODEL was used to analyze the 3D structure of motif 2, and the protein template used was (ID: 4oja.1) structure.The results show that motif 2 contains 4 β-sheets and 1 α-spin in the first and second. Between the β-sheets. The consistency of motif 2 with the template is 76.21 variant sites as shown in Figure 4.
 Figure 4 Sequence and mutation site of motif 2 |
1.3 SOD family evolutionary tree analysis of Maize
MEGA6.0 software was used to compare the similarity of SOD protein sequence, and adjacency method was used to draw evolutionary tree. There were two branches in the tree. ZmSOD1, ZmSOD3, ZmSOD4, ZmSOD5, ZmSOD7, ZmSOD8, ZmSOD10 and ZmSOD13 were branch I, ZmSOD2, ZmSOD12, ZmSOD14, ZmSOD9, ZmSOD11, ZmSOD6 and ZmSOD15 were branch II. Branches differ greatly from protein domains, which may be related to environmental factors. They may perform similar functions and need further experimental verification (Figure 5).
 Figure 5 SOD gene structure of Maize |
1.4 Comparison of SOD families among different species
The SOD conserved domain was used to search the database of sorghum and millet SOD gene families, and 18 SOD genes of sorghum and sorghum were selected. The corresponding protein sequences were compared with the maize SOD protein sequence to map the evolutionary tree. The genetic relationship shown in Figure 6 shows that the proteins with similar structures are clustered into one class. Each of the smallest branches has a maize sequence, a sorghum sequence and a gluten sequence. It can be seen that the SOD genes of these three species are highly homologous.
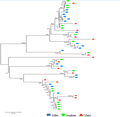 Figure 6 Cluster analysis of SOD protein in different species of Maize |
2 Discussion
With the in-depth study of plant genomics, a large number of plant genome sequences have been identified successively. The expression and identification of transcription factor genome has become a hotspot in the study of plant gene function. In recent years, SOD gene family members of different plant species have been analyzed,such as maize (Du et al., 2006), millet (Zhao et al., 2018), Arabidopsis (Guo et al., 2015). It has been found that transcription factors in SOD gene family can regulate multiple physiological processes in plants.
SOD proteins are classified into three types: SOD1, SOD2 and SOD3. SOD1 contains a barreled structure formed by beta helix and contains intramolecular disulfide bonds, and each subunit has a binuclear Cu/Zn site, which contains copper and zinc ions responsible for catalyzing superoxide disproportionation (Filiz and Tombuloglu, 2015). The gene encoding SOD2 usually contains five exons and four introns. This gene is located in the mitochondria. It encodes a homologous tetramer protein containing Fe/Mn locus in the mitochondrial matrix and forms a triangular bipyramidal geometric structure. The domain is formed by hydrogen bonding of amino acid residues. SOD3 belongs to extracellular domain (Wang et al., 2016). Superoxide dismutase and SOD3 are secreted to extracellular space and form glycosylated homologous tetramers, which can interact with heparan sulfate proteoglycan and collagen on cell surface and play a catalytic role (Yang et al., 2016).
SOD chromosome mapping found that maize contains 15 SOD gene family members, the relative number is small, the number of distribution on each chromosome is different, the gene structure is relatively simple, most of them encode hydrophilic proteins. These results indicate that these genes are relatively stable in structure, difficult to produce variable splicing when replicating, and relatively stable in function in plants.
In this study, bioinformatics was used to analyze SOD proteins in maize. According to their structures, SOD proteins in maize can be divided into two categories: Cu/Zn and Fe/Mn sequences, which are distributed at N and C ends, respectively. By mapping evolutionary trees, we found that there was only one SOD sequence in maize, millet and Sorghum at each branch of evolutionary trees, which indicated that SOD in plants of the same family and genus had strong structural similarity. This study also predicted 15 SOD proteins in maize. The results showed that the members of the same subfamily were similar in nature, indicating that the functions of gene family were relatively conservative and unchanged during evolution. They were called direct homologous genes. However, the physicochemical properties of the members of different subfamilies were poor. The reason is that the family genes form new biological functions in the process of evolution, and then produce side-line homologous genes to ensure that organisms are better adapted to the environment (Thornton and De, 2000). This is consistent with Chen Xuezhao's research conclusion (Chen, 2015).
3 Materials and Methods
3.1 Maize SOD family identification
The hidden Markov model of the SOD family (PF02777, PF00080, PF00081) was obtained from the Pfam online database (https://pfam.xfam.org/) (Punta et al., 2008). Gene ID, protein ID and protein sequences associated with the maize SOD gene family were searched using the Ensembl Plants online database. The redundant sequences were removed, resulting in a repeat-free gene, transcript and protein ID, protein sequence and number of exons. Protein structure detection was performed using CDD and SAMR online sites (Marchler-Bauer et al., 2009). Finally, the ProtParam online database was used to obtain information about SOD protein in maize, such as protein ID, chromosomal location, protein sequence, number of exons, molecular weight, number of amino acids, and isoelectric point.
3.2 Chromosome location analysis of SOD gene in Maize
The SOD gene database information in maize was used to find the length and location of maize chromosomes from Ensemble Plants. MapChatr software was used to draw genes and find relative positions on chromosomes. The physical location of genes on chromosomes was mapped (Wang et al., 2011).
3.3 Protein structure analysis of maize SOD family
The SOD protein sequence was analyzed by ProSite online website, and the corresponding domain (Sigrist et al., 2010) of each SOD protein was obtained. The conservative base library (Bailey et al., 2009) of corn CCT was obtained by online MEME tool. The three-dimensional structure of the obtained sequence was predicted by SWISS-MODEL.
3.4 Analysis of maize SOD phylogenetic tree
Using Clustal Omega comparative analysis, the SOD protein sequence of maize was obtained. The evolutionary tree was plotted by adjacency method using MEGA6.0 software, and bootstrap was set to 1000 (Kiefer et al., 2009).
3.5 Evolutionary analysis of SOD family among different species
SOD sequences in sorghum and millet were selected and screened according to 1.1 method. Redundancy was removed. SOD protein sequences of these two species were compared with those of maize. The results showed that the evolutionary tree was drawn by adjacency method using MEGA6.0 software, and bootstrap was set to 1000 (Tamura et al., 2013).
4 Conclusion
Fifteen SOD family members were identified by bioinformatics. Clustering analysis with millet, sorghum and SOD gene family, structural domain, gene structure and chromosome distribution, and other bioinformatics information showed that these genes were conservative in the evolutionary process, which could reveal the function of SOD protein family in Maize by genetic engineering in the future. It can provide a theoretical basis for the improvement of maize varieties.
Authors' contributions
GD carried out the molecular genetic studies, participated in the sequence alignment and drafted the manuscript. XHL, ZBY participated in the sequence alignment. MW, DZS, LCF participated in the design of the study and performed the statistical analysis. ZM conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Acknowledgments
The Key National Research and Development Program of China (2016YFD0300207, 2017YFD0301307)
Allen R.D., Webb R.P., and Schake S.A., 1997, Use of transgenic plants to study antioxidant defenses, Free Radical Biology Medicine, 23(3): 473-479
https://doi.org/10.1016/S0891-5849(97)00107-X
Bailey T.L., Boden M., Buske F.A., et al., 2009, MEME suite:Tools for motifdiscovery and searching, Nucl. Acids Res., 37(2): 202-208
https://doi.org/10.1093/nar/gkp335
PMid:19458158 PMCid:PMC2703892
Bannister J.V., Bannister W.H., and Rotilio G., 1987, Aspects ofthe structure, function, and applications of superoxide dis-mutas, Critical Reviews in Biochemistry and Molecular Bi-ology, 22(2): 111-180
https://doi.org/10.3109/10409238709083738
PMid:3315461
Baum J.A., and Scandalios J.G., 1981, Isolation and characteriza-tion of the cytosolic and mitochondrial superoxide dismu-tases of maize, Arch Biochem Biophys, 206(2): 249-264
https://doi.org/10.1016/0003-9861(81)90089-8
Bryan D.M., Julia M., and Kim S.J., 2000, Iron-superoxide dismutase expression in transgenic Alfalfa increases winter survival without a detectable increase in photosynthetic oxidative stress tolerance, Plant Physiology, 122: 1427-1437
https://doi.org/10.1104/pp.122.4.1427
PMid:10759538 PMCid:PMC58977
Chen S., Zhao Q.F., Chen H.L., and Jia L.Q., 2018, Bioinformatics and expression analysis of ubiquitin conjugating enzyme gene family in maize, Journal of South9ern Agriculture, 49(8):1476-1483
Chen X.Z., Zhang L., Yu S.S., Bi C.H., Liu C.Y., and Zhu Q., 2015, Molecular cloning and expression analysis of ubiquitin-conjugating enzyme E2-D2 in roughskin sculpin, Trachidermus Fasciatus, Acta Oceanologica Sinica, 37(10): 133-140
Droillard M.J., and Paulin A., 1990, Isozymes of superoxide dis-mutase in mitochondria and peroxisomes isolated frompetals of carnation (Dianthus caryophyllus) during senes-cence, Plant Physiol., 94(3): 1187-1192
https://doi.org/10.1104/pp.94.3.1187
PMid:16667815 PMCid:PMC1077360
Du J., Zhu Z., and Li W.C., 2006, Over-expression of Exotic superoxide dismutase gene MnSOD and increase in stress resistance in maize, Journal of Plant Physiology and Molecular Biology, (1): 57-63
Duke M.V., and Salin M.L., 1985, Purification and characterization of an iron-containing superoxide dismutase from a eucaryote, Ginkgo biloba, Arch Biochem Biophys, 243(1): 305-314
https://doi.org/10.1016/0003-9861(85)90800-8
Faize M., Burgos L., Faize L., Piqueras A., Nicolas E., Barba Espin G., Clemente-Moreno M.J., Alcobendas R., Artlip T., Her-nandez J.A., 2011, Involvement of cytosolic ascorbate peroxidase and Cu/Zn-superoxide dismutase for improved tolerance against drought stress, J. Exp. Bot., 62: 2599-2613
https://doi.org/10.1093/jxb/erq432
PMid:21239380
Feng C.J., Luo X.Y., Sha W., and Wang F.G., 2005, Effect of low temperature stress on SOD、POD activity and proline content of alfalfa, Pratacultural Science, 22(6): 29-32
Filiz E., and Tombuloglu H., 2015, Genome-wide distribution ofsuperoxide dismutase ( SOD) gene families in Sorghum bicolor, Turk. J. Biol., 39(1): 49-59
https://doi.org/10.3906/biy-1403-9
Guo C.M., Chen Z.M., and Chen C.L., 2015, AtADC2 enhances salt tolerance through regulating activities of superoxide dismutase and catalase in Arabidopsis thaliana, Plant Physiology Communications, 51(7): 1067-1074
Guo L.H., Wu X.L., and Gong M., 2005, Roles of glutathione reductase and superoxide dismutase in heat-shock-induced cross adaptation in maize seedlings, Plant Physiology Communications, 41(4): 429-432
Karuppanapandian T, Moon J-C, Kim C, Manoharan K, and Kim W., 2011, Reactive oxygen species in plants: their generation, signal transduction, and scavenging mechanisms, Aust J Crop Sci., 5(6): 709-725
. PDF(1009KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Dong Guo
. Mei Du
. Hanlin Xu
. Baoyuan Zhou
. Wei Ma
. Zaisong Ding
. Congfeng Li
. Ming Zhao
Related articles
. Zea mays
. Superoxide dismutase
. Bioinformatics analysis
Tools
. Email to a friend
. Post a comment


