2.Plant Breeding Division, Bangladesh Rice Research Institute, Gazipur, Bangladesh
3.Genetics and Plant Breeding Department, Bangladesh Agricultural University, Mymensingh, Bangladesh
BRAC
International Rice Research Institute
International Rice Research Institute
Bangladesh Agricultural University
 Author
Author  Correspondence author
Correspondence author
Molecular Plant Breeding, 2012, Vol. 3, No. 10 doi: 10.5376/mpb.2012.03.0010
Received: 15 Jun., 2012 Accepted: 25 Jun., 2012 Published: 29 Jun., 2012
Islam et al., 2012, Validation of SalTol Linked Markers and Haplotype Diversity on Chromosome 1 of Rice, Molecular Plant Breeding, Vol.3, No.10 103-114 (doi: 10.5376/mpb.2012.03.0010)
To determine the usefulness of SSR markers associated with a major QTL for salinity tolerance designated as "SalTol" collection of 115 diverse rice genotypes was phenotyped and genotyped using tightly linked DNA markers. These markers include five SSRs, RM1287, RM8094, RM3412, RM493 and RM140, and two EST markers, CP6224 and CP03970, on chromosome 1. Among the seven markers, the highest number of alleles (15) was found in RM8094, followed by 10 in RM187, RM3412 and RM493. The polymorphic information content (PIC) values ranged from 0.54 to 0.89. The highest PIC value (0.89) was found for RM8094, followed by RM493 and RM3412 (0.81) and RM1287 and RM140 (0.77). The marker RM8094 was useful for discriminating between tolerant and susceptible genotypes and therefore may be useful for marker-assisted selection. Seven haplotypes were identified among the 115 genotypes using the haplotype of IR66946-3R-178-1-1 (FL478) – the most widely used tolerant parent – which was used as a reference haplotype. Four genotypes had the same haplotype as FL478 and Pokkali-1. Of the seven different Pokkali seed sources tested, Pokkali-1 (IRGC 8948) contributed the SalTol region on the chromosome 1 segment of FL478. Genotypes from the haplotypes (in comparison with FL478) 1, 2 and 5 may be important for selecting alternative tolerant parents.
Rice (Oryza sativa L.) is a cereal plant of the grass family whose seeds are among the most important foods of the world and the most consumed staple in Asia. Recently, because of land scarcity and high food demand, to attain food security, marginal lands such as saline-prone areas must be put into use.
Salinity is an important abiotic problem in the rice-growing areas of the world. Salinity-affected areas have increased day by day because of the intrusion of brackish water during the dry season and at the start of the wet season and also because of the excessive use of irrigation water with improper drainage coupled with the use of poor-quality irrigation water (Ismail et al., 2010). Millions of hectares in the tropics and arid and semi-arid regions are technically suitable for rice cultivation. However, they are left idle or they grow very low yielding varieties due to the lack of suitable tolerant high-yielding modern varieties. Some 400-950 million hectares of global land are affected by different concentrations of salinity (Lin et al., 1998). The rice plant is one of the most suitable crops for saline soils, although it is considered moderately sensitive to salinity (Mori and Kinoshita, 1987).
Salt tolerance is a complex quantitative genetic character controlled by many genes (Shannon, 1985; Yeo and Flowers, 1986). Using conventional breeding methods, plant selection for salt tolerance is not easy because of the large effect of environment and low heritability of salt tolerance (Gregorio and Senadhira, 1993; Gregorio, 1997). Because of the complexity of the trait, it has been difficult to develop an accurate, rapid and reliable screening technique. Molecular markers are widely accepted as potentially valuable tools for crop improvement of rice (Mackill et al., 1999; McCouch and Doerge, 1995). Molecular markers will play a vital role in enhancing global food production by improving the efficiency of conventional plant breeding programs (Collard and Mackill, 2008). The progress and technical advances in molecular marker technology facilitate the identification of the major genes/QTLs for biotic and abiotic stresses in rice. The development of molecular marker selection permits the rapid and accurate identification of individuals that contain genes for salt tolerance.
DNA-based markers can be used to assess genetic diversity across an entire genome or specific chromosome regions. Bai et al (2003) and Liu and Anderson (2003) used microsatellite markers associated with a major QTL on chromosome 3BS for Fusarium head blight resistance identified in Sumai 3 to identify lines that putatively carry the 3BS QTL. To determine the suitability of identified markers linked to a QTL, SSR haplotyping was used, which has recently been used in rice. Diverse genotypes were selected to determine the level of polymorphism of SSR markers and to compare the haplotypes of tolerant or resistant genotypes.
An experiment with quantitative trait loci (QTL) for salt tolerance in rice was conducted using amplified fragment length polymorphism (AFLP), restriction fragment length polymorphism (RFLP) and microsatellite markers in different populations (Gregorio, 1997; Lang et al., 2000; Tuan et al., 2000; Bonilla et al., 2002; Niones, 2004; Thomson et al., 2010; Islam et al., 2011a). Microsatellite markers have been effective in mapping QTLs associated with salt tolerance (Lang et al., 2001). A major QTL for salt tolerance was mapped on chromosome 1 by using an F8 recombinant inbred line (RIL) of a Pokkali/IR29 cross (Gregorio, 1997). This QTL on chromosome 1 controlled the Na-K absorption ratio and accounted for 64.3 to 80.2% of the phenotypic variation in salt tolerance with LOD>14.5. This chromosome 1 segment was further saturated using RFLP and SSR markers using the RIL (Bonilla et al., 2002). The identified Na+, K+ and Na-K absorption ratio QTLs accounted for 39.2, 43.9 and 43.2% of the phenotypic variation with LOD>6.7. This segment of chromosome 1 was further fine mapped by using near isogenic lines (NILs) of the backcross of Pokkali/IR29 (Niones, 2004) with microsatellite markers. Pokkali has been the most widely used salt-tolerant parent by rice breeders. A newly developed elite line, FL478, from the cross of IR29 and Pokkali became popular as a novel source of salinity tolerance at seedling stage, because it possessed higher salinity tolerance than Pokkali and possessed many desirable traits.
The objectives of this study were to (i) compare the SSR marker haplotypes of salinity-tolerant genotypes with those of FL478 and Pokkali at the known SalTol QTL on chromosome 1; (ii) identify salinity-tolerant rice varieties/lines with putatively novel salinity tolerance sources; and (iii) identify the contributor of the SalTol region on chromosome 1 in FL478.
1 Results
The seedling stage salinity tolerance evaluation demonstrated that 37 genotypes were rated as tolerant to highly tolerant (score 1 and 3), 42 were moderately tolerant (score 5) and 36 were susceptible (score 7 and 9) (Table 1 and Table 3).
 Table 1 Origin, pedigree and salinity tolerance reaction of 115 rice varieties/lines |
Among the seven SalTol linked markers, the highest number of alleles was found in RM8094 (15), followed by RM1287, RM3412 and RM493 (10), RM140 (8), CP03970 (5) and CP6224 (3), which produced the lowest number of alleles, and the average was 8.71 (Table 2). The PIC values ranged from 0.54 to 0.89, with an average of 0.75. The highest PIC value (0.89) was found for the RM8094 locus followed by RM493 and RM3412 (0.81) and RM1287 and RM140 (0.77).
Table 2 Position of seven markers on chromosome 1 segment, their allelic variability, PIC values and amplicon size range for the SalTol QTL |
Only seven haplotypes were identified among the 115 genotypes (Figure 1) when the best three SSR markers were used to compare haplotypes using FL478 as a reference. The entries that had one of the FL478-type marker alleles for locus RM8094 were found to be either tolerant or moderately tolerant of salinity (Table 3). The varieties Pokkali-2, Pokkali-4, Pokkali-6, Kalimekri, Cheriviruppu, IR886-30-3-1-4-2, Bhirpala, CR1015, Kajalsail, IR71907-3R-2-1-2, IR71897-3R-1-1-2, IR55182-3B-14-3-2-3, IR58430-6B-14-1-2, IR63295-AC211-3, IR64197-3B-8-2, IR74099-3R-2-2, IR71991-3R-2-1, IR65192-4B-14-1, IR60483-2B-17-2-1-2, IR72048-B-R-11-1-3-1-2B-2, IR58443-6B-10-3 and IR71991-3R-2-6-1 had no common FL478 marker alleles for RM8094, RM3412 and RM493 (Table 3) but they were found to be highly tolerant of salinity (score 3) at EC 12 dSm-1.
Figure 1 Different haplotypes for FL478 |
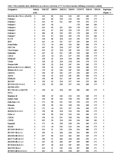 Table 3 Microsatellite allele distribution in a diverse collection of 115 rice lines/varieties differing in reaction to salinity |
Pokkali-1 possessed the same marker alleles as FL478 in the SalTol region on chromosome 1. Among the seven microsatellite markers, all the markers produced the same SSR marker alleles as FL478 except for RM140; this was similar to the other parent, IR29 (Table 4 and Figure 2).
Figure 2 Gel picture of five SSR markers of seven different Pokkali accessions, FL478 and IR29 |
Table 4 Modified standard evaluation score (SES) of visual salt injury at seedling stage |
2 Discussion
The purpose of this study was to test the linked microsatellite markers for the SalTol QTL on chromosome 1 in a diverse collection of salinity-tolerant and susceptible rice germplasm from different origins. This study illustrates the utility of microsatellite markers to identify the presence of the SalTol QTL in rice genotypes. From the phenotypic screening for salinity tolerance, it was found that, among the 115 genotypes, 37 were tolerant to highly tolerant (score 1 and 3), 42 were moderately tolerant (score 5) and 36 were susceptible (score 7 and 9) (Table 1 and Table 3). Most of the traditional varieties cultivated in saline-prone regions were found to be salt tolerant and most of the IRRI and BRRI-released modern high-yielding varieties (HYV) were found to be susceptible. Most of the 67 IRRI-developed elite lines from the salinity program were tolerant to moderately tolerant, indicating that these tolerant genotypes were derived from the active effort to breed and select salt-tolerant genotypes.
Based on the consensus map (www.tigr.org) of rice, these marker positions were from 10.9 Mb to 13.8 Mb or 58.1 cM to 66.5 cM (www.gramene.org) on the chromosome 1 segment where the SalTol QTL is located. Among the seven SSR markers analyzed, the highest number of alleles (15) was found in RM8094, followed by RM1287, RM3412 and RM493 (10), RM140 (8) and CP03970 (5), whereas CP6224 (3) produced the lowest number of alleles (Table 2). These results revealed that marker RM8094 would be useful and more information can be derived for screening rice germplasm for the SalTol QTL on chromosome 1, followed by RM1287, RM3412, RM493 and RM140. CP03970 and CP6224 could be used only for specific crosses. The PIC values for the seven markers are shown in Table 2, with a range from 0.54 to 0.89. The highest PIC value (0.89) was found for the RM8094 locus followed by RM493 and RM3412 (0.81) and RM1287 and RM140 (0.77). Therefore, RM8094, and to a lesser extent RM3412 and RM493, can be useful for marker-assisted selection of the SalTol QTL.
Based on marker analysis of Pokkali-1, IR29, FL478 and another RIL derived from the same IR29/Pokkali cross (FL378) and the phenotypes of these lines; we believe that the SalTol locus is located within the region of the three markers RM8094, RM3412 and RM493, as shown in Figure 2. This result was consistent with our previous mapping experiments, which found that RM8094 is the most tightly linked marker of the SalTol QTL, with the highest LOD peak (Islam et al., 2011b).
Seven haplotypes were identified among the 115 genotypes (Figure 1) when the marker alleles for the best three SSR markers were used to compare haplotypes using FL478 as a reference. The FL478 haplotype (haplotype 1) was found in five genotypes, and this was very important for tracking SalTol with molecular markers. Among the 115 genotypes, 76 genotypes had none of the same marker alleles as the FL478 haplotype (haplotype 7).
Among the 12 popular BRRI-developed rice varieties, all had haplotypes different from that of FL478. Different marker alleles from FL478 or Pokkali were produced for the marker RM8094 in the BRRI varieties.
Considering the haplotypes for FL478, it was found that the entries that had one of the FL478-type marker alleles for locus RM8094 were found to be either tolerant or moderately tolerant of salinity (Table 3). It was found that lines PSBRc82, IR4630-22-2-5-1-5, Pelita I-1, IR66404-4B-20-1-2, IR71866-3R-1-2-1, IR65775-4B-19-1-3 and PSBRc50 had the FL478 marker allele for the RM8094 marker and all of these lines/varieties were found to be tolerant to moderately tolerant of salinity (score 3 and 5); none of them were susceptible (Table 3). Therefore, marker RM8094 appears to be diagnostic of salinity tolerance. The 115 genotypes that had the same allele as FL478 for marker RM3412 were found to have a mixed reaction to salinity stress (Table 3). A similar result was also found for marker locus RM493. Therefore, these two markers (RM3412 and RM493) must not be used for MAS in populations derived from these susceptible parents.
It was also found that some of the varieties, such as Pokkali-2, Pokkali-4, Pokkali-6, Kalimekri, Cheriviruppu, IR886-30-3-1-4-2, Bhirpala, CR1015, Kajalsail, IR71907-3R-2-1-2, IR71897-3R-1-1-2, IR55182-3B-14-3-2-3, IR58430-6B-14-1-2, IR63295-AC211-3, IR64197-3B-8-2, IR74099-3R-2-2, IR71991-3R-2-1, IR65192-4B-14-1, IR60483-2B-17-2-1-2, IR72048-B-R-11-1-3-1-2B-2, IR58443-6B-10-3 and IR71991-3-R-2-6-1, had no common FL478 marker alleles for RM8094, RM3412 and RM493 (Table 3). However, these lines were highly tolerant of salinity (score 3) at EC 12 dS m-1. These results suggest that QTLs other than SalTol control salinity tolerance in these tolerant genotypes. Therefore, these genotypes may represent novel sources of salinity tolerance, and could potentially be exploited to identify new QTLs other than SalTol.
For the seven different sources of Pokkali that were tested, we found that all of them were different except for Pokkali-5 and Pokkali-7. The most widely used salt-tolerant line, FL478, was derived from the cross between IR29 and Pokkali. However, the original Pokkali accession used for making this cross was not properly documented. So, in this study, we were trying to identify which source of Pokkali contributed the SalTol region in FL478. Results showed that Pokkali-1 possessed the same marker alleles as FL478 in the SalTol region in FL478. Among the seven microsatellite markers, all the markers produced the same SSR marker alleles as FL478 except for RM140; this was similar to the other parent, IR29 (Table 3 and Figure 2). This result suggested that the region containing the locus RM140 was contributed by IR29 and other segments containing the marker loci RM8094, RM3412 and RM493 by Pokkali-1. Therefore, it was suggested that, among the seven Pokkali sources, Pokkali-1 contributed the SalTol region in FL478. This result was contrary to the previous report of Walia et al (2005), who reported that IR29 was the source of the SalTol region in FL478.
The SSR markers linked to the SalTol QTL were analyzed and showed that RM8094, RM3412 and RM493 are useful markers for identifying genotypes with salinity tolerance at the seedling stage. These validated SSR markers can be used for marker-assisted selection to introgress the SalTol QTL from a donor to a recurrent genotype. The FL478 haplotype was found in five genotypes, Pokkali-1, FL378, IR4630-22-2-5-1-5, Pelita I-1 and PSBRc50, which were very important for tracking SalTol with molecular markers. The genotypes Kalimekri, Cheriviruppu, IR886-30-3-1-4-2, Bhirpala, CR1015, Kajalsail, IR71907-3R-2-1-2, IR71897-3R-1-1-2, IR55182-3B-14-3-2-3, IR58430-6B-14-1-2, IR63295-AC211-3, IR64197-3B-8-2, IR74099-3R-2-2, IR71991-3R-2-1, IR65192-4B-14-1, IR60483-2B-17-2-1-2, IR72048-B-R-11-1-3-1-2B-2, IR58443-6B-10-3 and IR71991-3R-2-6-1 were identified as potential new sources of salinity tolerance other than SalTol. These new findings can redirect breeding strategies for salinity-tolerant rice to develop a new generation of salinity-tolerant varieties.
3 Materials and Methods
3.1 Phenotyping of diverse rice genotypes
The usefulness of the identified molecular markers linked to the SalTol QTL on chromosome 1 can be confirmed when applied on a wider scale to 115 diverse rice genotypes (Table 1). These genotypes were composed of 67 IRRI-developed elite lines for salt tolerance, 18 IRRI-released modern varieties and 12 BRRI (Bangladesh Rice Research Institute)-released modern varieties, along with 3 traditional Bangladeshi varieties cultivated in coastal regions, 9 varieties of Indian origin, 2 varieties from both Vietnam and Indonesia, one variety from Pakistan and one from the Philippines. We used 7 Pokkali accessions: Pokkali-1 (Acc. # IRGC8948), Pokkali-2 (Acc. # IRGC15238), Pokkali-3 (Acc. # IRGC15388), Pokkali-4 (Acc. # IRGC15602), Pokkali-5 (Acc. # IRGC15661), Pokkali-6 (Acc. # IRGC108921) and Pokkali-7 (Acc. # IRGC15661) to identify the contributor of SalTol in FL478 with other genotypes. All seeds were collected from the IRRI Plant Breeding, Genetics, and Biotechnology Division.
We screened 115 genetically diverse rice genotypes under controlled environmental conditions in the phytotron at IRRI with IR29 and FL478 as susceptible and tolerant checks, respectively. Twenty pregerminated seeds per genotype were transplanted in 100-hole seedling floats. Seedling stage tolerance screening was set up (Gregorio et al., 1997) using nutrient solution described by Yoshida et al (1976). A salinity of EC 12 dSm-1 was applied 4 days after transplanting. The screening was conducted in the IRRI phytotron with temperature maintained at 29℃/21℃ day/night, relative humidity of 70% during the day and natural daylight. A modified standard evaluation score (Table 4) was used in rating the symptoms of salt damage (Gregorio, 1997). Ratings of salt injury symptoms were recorded 3 weeks after salinization when the susceptible check IR29 was dead.
3.2 Molecular marker analysis
Previous reports (Islam et al., 2005; Islam et al., 2011a) found that four SSR markers and one EST marker were linked to the SalTol QTL on the chromosome 1 segment. Niones (2004) also reported one more SSR and EST marker linked to this QTL. So, a total of five SSR markers and two EST markers were used in this study.
Genomic DNA of the 115 genotypes was extracted using the CTAB method described by Zheng et al (1995). PCR was performed following the protocol described by Temnykh et al (2000) using a PTC-100 dyad thermocycler machine (M J Research) with 384-well plates. Amplification products were resolved by 8% polyacrylamide gel electrophoresis. Gels were run for 2~3.5 hours at 100 volts and stained in ethidium bromide and visualized under UV light.
The molecular weight for each SSR marker allele was measured by using Alfa Imager software version 5.5. Polymorphic information content (PIC) values of the SSR markers were calculated according to the formula of Anderson et al (1993). SSR marker alleles were analyzed using the program Power Marker version 3.25 (Liu and Muse, 2005). Haplotype analysis was conducted according to Bai et al (2003) and Liu and Anderson (2003) using the best three SSR markers, i.e., RM8094, RM3412 and RM493, to compare with FL478 as a reference.
Acknowledgments
We thank Poverty Elimination through Rice Research Assistance (PETRRA), the Generation Challenge Program (GCP) and Challenge Program for Water and Food (CPWF) for providing funds for doing this research. We also thank BRRI and IRRI for allowing us to do this research and for technical support.
References
Anderson J.A., Churchill G.A., Autrique J.E., Tanksley S.D. and Sorrells M.E., 1993, Optimizing parental selection for genetic linkage maps, Genome, 36: 181-186
http://dx.doi.org/10.1139/g93-024 PMid:18469981
Bai G., Peiguo G., and Kolb F.L., 2003, Genetic relationship among head blight resistant cultivars of wheat assessed on the basis of molecular markers, Crop. Sci., 43: 498-507
http://dx.doi.org/10.2135/cropsci2003.0498
Bonilla P.S., Dvorak J., Mackill D.J., Deal K., and Gregorio G., 2002, RFLP and SSLP mapping of salinity tolerance genes in chromosome 1 of rice (Oryza sativa L.) using recombinant inbred lines, Philipp Agric. Sci., 85(1): 64-74
Collard B.C.Y., and Mackill D.J., 2008, Marker-assisted selection: an approach for precision plant breeding in the twenty-first century, Phil. Trans. Soc. B., 363: 557-572
http://dx.doi.org/10.1098/rstb.2007.2170 PMid:17715053 PMCid:2610170
Gregorio G.B. and Senadhira D., 1993, Genetics analysis of salinity tolerance in rice, Theor. Appl. Genet., 86:333-338
http://dx.doi.org/10.1007/BF00222098
Gregorio G.B., 1997, Tagging salinity tolerance genes in rice using amplified fragment length polymorphism (AFLP), Ph.D. dissertation, College, Laguna, Philippines: University of the Philippines Los Baños, Laguna, pp.118
Gregorio G.B., Senadhira D., and Mendoza R.D., 1997, Screening rice for salinity tolerance. IRRI discussion paper series no. 22. IRRI, DAPO Box 7777, Metro Manila, Philippines. pp.3-19
IRRI (International Rice Research Institute), 1997, Annual Report for 1997, Manila (Philippines): International Rice Research Institute, pp.308
Islam M.R., Gregorio G.B., Salam M.A., Collard B.C.Y., Tumimbang-Raiz E., Adorada D.L., Mendoza R.D., Singh R.K., and Hassan L., 2011(b), Validation of a major QTL for salinity tolerance on chromosome 1 of rice in three different breeding populations, Agrochimica, 55(6): 355-366
Islam M.R., Gregorio G.B., Salam M.A., Hassan L., Tumimbang-Raiz E., Adorada D.L. and Mendoza R.D., 2005, Molecular markers to salt tolerance in rice: Is it working? In: Abstracts, 5th International Rice Genetics Symposium, IRRI, Manila, Philippines, pp.115-116
Islam M.R., Salam M.A., Hassan L., Collard B.C.Y., Singh R.K., and Gregorio G.B., 2011(a), QTL mapping for salinity tolerance at seedling stage in rice, Emirates J. Food. Agric., 23 (2): 137-146
Ismail A.M., Thomson M.J., Vergara G.V., Rahman M.A., Singh R.K., and Gregorio G.B., 2010, Designing resilient rice varieties for coastal deltas using modern breeding tools. In: Tropical deltas and coastal zones: food production, communities and environment at the land-water interface, Wallingford: CABI, pp.154-165
http://dx.doi.org/10.1079/9781845936181.0154
Lang N.T., Li. Z.K., and Bui C.B., 2001, Microsatellite markers linked to salt tolerance in rice, Omonrice, 9: 9-21
Lang N.T., Yanagihara S., and Buu B.C., 2000, Quantitative trait loci for salt tolerance in rice via molecular markers, Omonrice, 8: 37-48
Lin H.X., Yanagihara S., Zhuang J.Y., Senboku T., Zheng K.L., and Yashima S., 1998, Identification of QTL for salt tolerance in rice via molecular markers, Chinese J. Rice Sci., 12(2): 72-78
Liu K., and Muse S.V., 2005, PowerMarker: integrated analysis environment for genetic marker data. Bioinformatics, 21: 2128-2129
http://dx.doi.org/10.1093/bioinformatics/bti282 PMid:15705655
Liu S., and Anderson J.A., 2003, Targeted molecular mapping of a major wheat QTL for Fusarium head blight resistance using wheat ESTs and synteny with rice, Genome, 46: 817-823
http://dx.doi.org/10.1139/g03-066 PMid:14608398
Mackill D.J., Nguyen H.T., and Zhan J., 1999, Use of molecular markers in plant improvement programs for rainfed lowland rice, Field Crops Res., 64: 177-185
http://dx.doi.org/10.1016/S0378-4290(99)00058-1
McCouch S.R., and Doerge R.W., 1995, QTL mapping in rice, Trends. Genet., 11: 482-487
http://dx.doi.org/10.1016/S0168-9525(00)89157-X
Mori I., and Kinoshita T., 1987, Salt tolerance of rice callus clones, Rice. Genet. Newsl., 4: 112-113
Niones J.M., 2004, Fine mapping of the salinity tolerance gene on chromosome 1 of rice (Oryza sativa L.) using near isogenic lines, MS dissertation, College, Laguna, Philippines: University of the Philippines Los Baños, Laguna
Shannon M.C., 1985. Principles and strategies in breeding for higher salt tolerance, Plant Soil, 89: 227-241
http://dx.doi.org/10.1007/BF02182244
Temnykh S., Park W.D., Ayres N., Carthinour S., Hauck N., lipovich L., Cho Y.G., Ishii T., and McCouch S.R., 2000, Mapping and genome organization of microsatellite sequences in rice (Oryza sativa L.), Theor. Appl. Genet., 100: 697-712
http://dx.doi.org/10.1007/s001220051342
Thomson M.J., Ocampo M., de Egdane J., Rahman M.A., Sajise A.G., Adorada D.L., Tumimbang-Raiz E., Blumwald E., Serraj Z.I., Singh R.K., Gregorio G.B., and Ismail A.M., 2010, Characterizing the Saltol quantitative locus for salinity tolerance in rice, Rice, 3: 148-160
http://dx.doi.org/10.1007/s12284-010-9053-8
Tuan V.D., Fukuta Y., Tano M., and Ban T., 2000, Mapping quantitative trait loci for salinity tolerance in rice, Omonrice, 8: 27-35
Walia H., Wilson C., Condamine P., Liu X., Ismail A.M., Zeng L., Wanamaker S.I., Mandal J., Xu J., Cui X., and Close T.J., 2005, Comparative transcriptional profiling of two contrasting rice genotypes under salinity stress during the vegetative growth stage. Plant Physiol, 139: 822-835
http://dx.doi.org/10.1104/pp.105.065961 PMid:16183841 PMCid:1255998
Yeo A.R and Flowers T.J., 1986, The physiology of salinity tolerance in rice (O. sativa) and a pyramiding approach to breeding varieties for saline soils, Aust. J. Plant Physiol., 13: 75-91
http://dx.doi.org/10.1071/PP9860161
Yoshida S., Forno D.A., Cock J.H. and Gomez K.A., 1976, Laboratory manual for physiological studies of rice, International Rice Research Institute (IRRI), Los Baños, Philippines
Zheng K., Huang N., Bennett J., and Khush G.S., 1995, PCR-based marker assisted selection in rice breeding, IRRI, Manila, Philippines, pp.16-18
. PDF(195KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Mohammad Rafiqul Islam
. Glenn Borja Gregorio
. Md. Abdus Salam
. Bertrand C. Y. Collard
. Rakesh Kumar Singh
. Lutful Hassan
Related articles
. Haplotype
. Rice
. Salinity tolerance
. Chromosome 1
Tools
. Email to a friend
. Post a comment


