Locus R-D1 Conferring Red-Grain-Color in Synthetic Derivative Wheat Chuanmai 42 Mapped with SSR Markers 
2. Institute of Plant Protection, Sichuan Academy of Agricultural Sciences, Chengdu, 610066, P.R. China
3. Institute of Biodiversity Science, Fudan University, Shanghai, 200433, P.R. China
 Author
Author  Correspondence author
Correspondence author
Molecular Plant Breeding, 2010, Vol. 1, No. 3 doi: 10.5376/mpb.2010.01.0003
Received: 06 Apr., 2010 Accepted: 29 Jun., 2010 Published: 28 Sep., 2010
Li et al., 2010, Locus R-D1 conferring red-grain-color in synthetic derivative wheat chuanmai 42 mapped with SSR markers, Molecular Plant Breeding Vol.1 No.3 (doi:10.5376/mpb.2010.01.0003)
Synthetic hexaploid wheat (SHW) (Triticum durum×Aegilops tauschii) holds a large number of unfavorable traits, of which, red grain color was important to limit the wheat breeder using SHWs for common wheat improvement. The objectives of this study were to identify and map the red grain color gene derived from SHW in a Chinese wheat variety Chuanmai 42 by SSR molecular markers. 1015 individuals in F2 population derived from the cross of Chuanmai 42 with a red grain color and ChuanW565 (CW565) with white grain color were used for tagging the gene of red grain color. A total of 40 SSR markers on the chromosome 3D were employed to detect the polymorphism of between the parents, Chuanmai 42 and CW565. 241 recessive individuals in F2 population derived from the cross (Chuanmai 42×CW565) were employed for allelic test of the red-grain gene by eight polymorphic markers. The results indicated that the red-grain color gene of Chuanmai 42 derived from the D genome donor of synthetic hexaploid wheat and controlled by a single dominant gene R-D1 on the long arm chromosome 3D within an interval of 4.6 flanked by SSR markers Xgwm3 and Xgwm314, and genetic distance were 2.1 cM, 2.5 cM, respectively. Those two SSR markers can be used in marker-assisted selection for developing new white grain wheat variety based on the progenies of SHWs or Chuanmai 42 crossed with white grain color wheat in wheat breeding program.
Background
Grain color is one of important traits affecting flour yield and quality in wheat. Red grain color of wheat commonly associated with the development of grain dormancy and affected brightness of wheat flour due to contamination of red pigment in milling process (Flintham, 2000; Warner et al., 2000; Himi et al., 2002). The most of wheat famers in the world prefer to grow white grain wheat varieties instead of red one for the wheat milling quality. Wheat grain color was controlled by the red seed color genes on the end region of the long arms of wheat chromosomes 3A, 3B and 3D, respectively (McIntosh et al., 1998). Each red grain color gene (R-A1 on 3A, R-B1 on 3B and R-D1 on 3D,) was dominant and inherited monoxenically. The red alleles were assigned as R-A1b, R-B1b and R-D1b, and the white alleles were assigned as R-A1a, R-B1a, and R-D1a (McIntosh et al., 1998), a single locus containing the dominant allele was sufficient to result in red color. The degree of red color was additive, the intensity of the red color depended on the number of R alleles, and only those homozygous recessive at all three genes being white (R-A1a, R-B1a and R-D1a).
Synthetic hexaploid wheat (SHW) (Triticum durum×Aegilops tauschii) was created to explore novel genes in T. durum and Ae. tauschii for common wheat improvement. A plenty of useful traits have been found in SHW such as disease resistance (Ma et al., 1995; Kema et al., 1995; Yang et al., 1999), pest resistance (Eastwood et al., 1991; Thompson et al., 1999; Hollenhorst and Joppa, 1983; Tyler and Hatchett, 1983), improved zinc efficiency (Cakmak et al., 1999; Genc and McDonald, 2004), salt tolerance (Gorham, 1990), cold tolerance (Limin and Fowler, 1993), water logging tolerance (Villareal et al., 2001), pre-harvest sprouting (Trethowan et al., 1998; Gatford et al., 2002), increased level of iron concentration in grain (Lage and Trethowan, 2008), end-use quality (William et al., 1993; Peña et al., 1995), and yield components (Villareal et al., 1994a; 1994b; Lage et al., 2006). However, SHWs also carried a large number of unfavorable traits, such as late maturity, tallness, difficulty in threshing and red grain color. The last one, red grain color, was an important factor limiting wheat improvement. Over one thousand SHWs have been produced from more than 600 Aegelops tauschii accessions by the International Maize and Wheat Improvement Center (CIMMYT) (Mujeeb et al., 1996; Hajjar and Hodgkin, 2007; Maarten et al., 2007).
To enhance the genetic diversity of Chinese wheat varieties, over two hundred synthetic hexaploid wheat accessions from CIMMYT were introduced into China. Elite synthetics were crossed and backcrossed with Chinese commercial wheat cultivars to improve stripe rust resistance, quality and yield potential. Advanced lines with good resistance and high yield potential were developed. In recent years, four synthetic derivatives have been released in China. Among them, Chuanmai 42, the first released CIMMYT synthetic derivative in the world with beneficial traits as large kernels, high spike weight, and resistance to new races of local stripe rust, had the highest average yield (>6t/ha) over two years in Sichuan provincial yield trials, outyielding the commercial check cultivar Chuanmai 107 by 20%~35% (Zhang et al., 2004; Hajjar and Hodgkin, 2007; Maarten et al., 2007). Now, Chuanmai 42 has become a popular wheat cultivar throughout southwestern China and is playing a significant role in wheat high yield breeding. However, the character of red grain color is a limiting factor that influences its distribution in some wheat region.
It was a cumbersome and time-intensive task to make a conversion from red-seeded type to white-seeded type, due to red grain color was dominant and three homozygous recessive loci were needed for the white-seeded type. Molecular markers have the potential to facilitate the effectiveness of the recessive allele selection for white variety breeding. Thus, for this project, we mapped the gene(s) controlling the character of red grain color in Chuanmai
42 derived from SHW by SSR markers, and provided useful markers for breeding white grain color variety by using SHWs and Chuanmai 42 as genetic resources.
1 Results
1.1 Inheritance of red grain color in Chuanmai 42
The F1 seeds of all the other crossed combination with Chuanmai 42 (red-grain color)×CW565 (white-grain color) were uniformly of red-grain color as Chuanmai 42, which confirmed that the red-grain color gene in Chuanmai 42 was dominant gene to white. Phenotypic analysis of F2 seeds found 774 red individuals and 241 white out of 1015 screened, which is not significantly different from the 3:1 ratio (x2=0.85, p>0.3) (Table 1), indicating that the character of red grain color in Chuanmai 42 was controlled by a single dominant gene.
Table 1 Segregation for red and white grain color in F2 progenies |
1.2 Linkage analysis and R-D1 mapping
According to the pedigree of Chuanmai 42, it was speculated that the red seed color alleles conferring Chuanmai 42 are R-A1a, R-B1a and R-D1b. Therefore, a total of 40 SSR markers on the chromosomes 3D were used to screen the polymorphisms between the two parents Chuanmai 42 and CW565. And eight SSR markers were observed polymorphic on the long arm of chromosome 3D.
The confirmed 241 recessive plants of F2 population were used to construct the linkage map of the gene of red grain color; the genetic distance between R-D1 gene and SSR loci was analyzed by Mapmaker 3.0b. Eight polymorphic SSR markers were employed to screen the 241 recessive plants, and five SSR markers (Xgwm3, Xgwm314, Xgwm664, Xwmc552 and Xbarc270) showed close linkage to R-D1 gene (Table 2, Figure 1). The R-D1 gene in Chuanmai 42 linked closely with markers Xgwm3 and Xgwm314 with 2.1 cM and 2.5 cM genetic distance, respectively (Figure 1).
Table 2 Linkage analysis of Red-grain-color gene R-D1 with five polymorphic SSR markers in 241 recessive plants of F2 population |
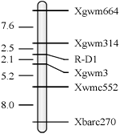 Figure 1 Linkage map involving the R-D1 gene constructed with five SSR markers on chromosome 3DL |
2 Discussion
Chuanmai 42, a wheat cultivar with high-yield potential, was developed by using a red grain color synthetic hexaploid wheat (Syn769, durum wheat Decoy1 (white grain color) ×Ae.tauschii SQ-188 (red grain color)) crossed and backcrossed with two white grain color local wheat varieties. According to the pedigree, the character of red grain color of Chuanmai 42 derived from the D genome donor Ae.tauschii SQ-188 of Syn769. The red grain color was dominant to white, and the alleles conferring red grain color were denoted as R-A1b, R-B1b and R-D1b,and alleles conferring white grain color as R-A1a, R-B1a and R-D1a. The character of red grain color in Chuanmai 42 was controlled by a single gene, so the red grain color alleles conferring Chuanmai42 were R-A1a, R-B1a and R-D1b. Chuanmai42 is becoming a popular cultivar and an important breeding parent in China because of a high and stable yield potential, despite having a red grain color while white is preferred.
To date, R-A1, R-B1 and R-D1 were mapped by RFLP, STS and EST marker. Previous mapping efforts showed that R genes were mapped by orthologous alleles of RFLP markers on the long arm of chromosome 3D, 3A and 3B (Flintham and Humphrey, 1993; Gale et al., 1995; Nalam et al., 2006; Kuraparthy et al., 2008), and R-D1 gene closely linked with RFLP markers Xbcd131 and Xabc174 on the long arm of chromosome 3D. R-A1 was mapped within an interval of 4.4 cM flanked by one STS marker and one EST marker (Kuraparthy et al., 2008). R-A1, R-B1 and R-D1 were also mapped by SSR (Sherman et al., 2008; Imtiaz et al., 2008; Groos et al., 2002), and R-D1 gene tightly linked to SSR markers Xgwm4306 (a distance of 3.2 cM) (Sherman et al., 2008) and Xgwm1200 (a distance of 5.0 cM) (Imtiaz et al.,2008). In present paper, the R-D1 gene derived from D genome donor of synthetic hexaploid wheat was also successfully mapped on the long arm of chromosome 3D within an interval of 4.6 cM flanked by SSR markers Xgwm3 and Xgwm314, with genetic distance of 2.1 cM and 2.5 cM, respectively. The SSR markers to R-D1 (Xgwm3 and Xgwm314) in our population were more tightly linked than reported previously.
Since 2006, we began to transfer the white grain color gene R-D1a of CW565 into Chuanmai 42 by selecting the heterozygous genotype on R-D1 allele with SSR markers Xgwm3 and Xgwm314 in the backcross progeny, and several advanced white grained lines with good agronomic traits as Chuanmai 42 were developed. The results indicated that the two SSR markers can be used in marker-assisted selection for breeding white grain wheat variety by discarding the red grain color gene or selecting the white grain color gene in the progeny of SHWs or Chuanmai 42 crossed with white grain color wheat.
3 Materials and methods
3.1 Plant materials
Two wheat lines, Chuanmai 42 with red grain color and ChuanW565 (CW565) (R-A1a, R-B1a and R-D1a) with white grain color were employed in the present experiment. Both Chuanmai 42 and CW565 were developed and provided by Crop Research Institute, Sichuan Academy of Agricultural Sciences in Sichuan province from China.
Chuanmai42 was developed using a red grain color synthetic hexaploid wheat (Syn769) crossed and backcrossed with two white grain color local wheat varieties Sw3243 (R-A1a, R-B1a and R-D1a) and Chuan6415 (R-A1a, R-B1a and R-D1a), respectively. The synthetic hexaploid wheat Syn769 was developed from a white grain color durum wheat Decoy1 (2n=4x=28, AABB) (R-A1a and R-B1a) and the red grain color Ae. tauschii accession SQ-188 (2n=2x=14, DD)(R-D1b) by CIMMYT. According to the pedigree, the character of red grain color of Chuanmai42 should be derived from the D genome donor SQ-188 of Syn769, and the red grain color alleles conferring Chuanmai42 are R-A1a, R-B1a and R-D1b.
The Chuanmai 42 was hybridized with CW565 under field conditions in Chengdu city of Sichuan province in 2004. For seed multiplication during the summer season, the F1 seeds were planted at Yunnan Agricultural University in Kunming city. During the winter season, the F2 populations, obtained from Yunnan Agricultural University, together with their parents and F1’s were grown as individual plants for analysis in the field of Chengdu city. A total of 1 015 F2 plants were harvested individually for estimating the grain color and analyzing the inheritance of the character of red color of Chuanmai 42 in the spring of 2005. For confirming the recessive plants, the 241 recessive lines of F2 population with white grain color were selected and planted (30~40 plants each) during the winter season. In the spring of 2006, the F2-3 single plants derived from 241 recessive lines were harvested and used to estimate the grain color again.
The 1 015 plants of F2 population of Chuanmai 42×CW565 was used for the inheritance of the red grain trait, and 241 F2 recessive lines confirmed by it’s F2-3 populations were used for the mapping of red grain color gene.
3.2 Trait evaluation
At the completely mature time, pericarp color of single plant F2 populations and F2-3 populations derived from the recessive lines of Chuanmai42×CW565, which was determined in grain color after single plant threshing. The grain color was evaluated by using NaOH method and visual assessments. For the NaOH method, thirty to forty seeds of each line were placed into 100 x 15 mm petri-dishes. A 5% NaOH solution was poured over the seeds and they were soaked in solution overnight. The NaOH solution gave red wheat a dark red color, while white wheat assumes a straw yellow color.
3.3 DNA extraction and microsatellite analysis
Total genomic DNA of the parents and F2 single plants was extracted from leaf material of individual plants using the CTAB protocol (Sharp et al., 1988). According to results of estimation of grain color on the F2 and F2-3 plants, the genomic DNA of 241 recessive lines were selected and used for molecular mapping. In all, 40 pairs of wheat SSR primers on chromosome 3D were used to screen on the two parents Chuanmai42 and CW565 of mapping population. Primer sequences were described by Roder et al. (1998), Pestsova et al. (2000), Daryl et al. (2004) and Song et al. (2005), and then synthesized by Shanghai Sangon Biological Engineering Technology & Services Co. Ltd (http: // www.sangon.com).
The PCR reaction was performed in a volume of 15 µL in a PTC100 Peltier Thermal Cycler. The reaction mixture contained 1.5 µL 10x buffer (50 mmol KCl, 10 mmol Tris-HCl pH 8.3), 1.5 mmol/L MgCl2, 200 µmol/L each dNTP, 250 nmol/L each primer, 1 unit Taq polymerase and 50~100 ng template DNA. The reagents in PCR reaction were obtained by Chengdu Feike BioTechnology Co., Ltd (http://www.fkbio.com). The PCR conditions were as follows: denaturation at 94℃ for 4 min, followed by 40 cycles of 94℃ for 1 min, 50~60℃ (depending on the individual primer) for 1 min, 72℃ for 1 min and a final extension at 72℃ for 10 min. PCR products were mixed with 6 µL formamide loading buffer (98% formamide, 10 mmol/LEDTA, 0.25% bromophenol blue, 0.25% xylene cynol, pH 8.0). Each sample of 8~10 µL was loaded on 6% denaturing polyacrylamide gels and run at 400 V for approximately 1 h and then dyied by the silver staining method as described by Bassam (1991).
3.4 Linkage analysis
The linkage map was generated using MAPMAKER/Exp version 3.0b (Lincoln et al., 1992). An LOD score of 3.0 was used as a threshold for the declaration of linkage, Genetic distances in centimorgans (cM) were calculated by applying the Kosambi (Kosambi, 1944) mapping function and. The genetic map was drawn with the software Mapdraw V2.1 (Liu and Meng, 2003).
Authors’ contributions
JL conducted the major part of this study including experimental design, construction of the F2 population, SSR analysis, and manuscript preparation. HTW participated SSR analysis and genetic linkage mapping. XRH participated construction of the F2 population. BRL assisted in the development of the project and manuscript preparation. WYY participated in the development of the project, experimental design and manuscript preparation. All authors read and approved the final manuscript.
Acknowledgements
The authors thank Jie Liao for her part experiments. This work was supported by the National 863 program (No. 2006AA10Z1C6), and the National Natural Science Foundation of China (No.30771338 and 30700495).
References
Bassam B.J., Caetano-Anolles G., and Gresshoff P.M., 1991, Fast and sensitive silver staining of DNA in polyacrylamide gels, Anal. Biochem., 196: 80-83 doi:10.1016/0003-2697(91)90120-I
Cakmak I., Cakmak O., Eker S., Ozdemir A., Watanabe N., and Braun H.J., 1999, Expression of high zinc efficiency of Aegilops tauschii and Triticum monococcum in synthetic hexaploid wheats, Plant and Soil, 215: 203-209 doi:10.1023/A:1004504726214
Daryl J.S, Isaac P., and Edwards K., 2004, A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.), Theoretical and Applied Genetics, 109: 1105-1114 doi:10.1007/s00122-004-1740-7
Eastwood R.F., Lagudah E.S., Appels R., Hannah M., and Kollmorgen J.F., 1991, Triticum tauschii: a novel source of resistance to cereal cyst nematode (Heterodera avenae), Australian Journal of Agricutural Research, 42: 69-77
Flintham J.E., and Humphrey S.J., 1993, Red coat genes and wheat dormancy, In: Kettlewell P.S. (eds.), Cereal quality III, Aspects of Applied Biology, Vol.36, Association of Applied Biologists, Wellesbourne, Warwick, UK. pp. 135-141
Flintham J.E, 2000, Different genetic components control coat imposed and embryo-imposed dormancy in wheat, Seed Science Research, 10: 43-50
Gale M.D., Atkinson M.D., Chinoy C.N., Harcourt R.L., Jia J., Li Q.Y., and Devos K.M., 1995, Genetic maps of hexaploid wheat, Proceedings 8th International Wheat Genetics Symposium (Li Z.S. and Xin Z.Y. eds.), China Agricultural Scientech Press, Beijing, pp.29-40
Gatford K.T., Hearnden P., Ogbonnaya F., Eastwood R.F., and Halloran G.M., 2002, Novel resistance to pre-harvest sprouting in Australian wheat from the wild relative Triticum tauschii, Euphytica, 126: 67-76 doi:10.1023/A:1019611403701
Genc Y., and McDonald G.K., 2004, The potential of synthetic hexaploid wheats to improve zinc efficiency in modern bread wheat, Plant and Soil, 262: 23-32 doi:10.1023/B:PLSO.0000037024.55764.26
Gorham J., 1990, Salt tolerance in the Triticeae: K/Na discrimination in synthetic hexaploid wheats, Journal of Experimental Botany, 41: 623-627 doi:10.1093/jxb/41.5.623
Groos C., Gay G., Perretant M.R., Gervais L., Bernard M., Dedryver F., and Charmet G., 2002, Study of the relationship between pre-harvest sprouting and grain color by quantitative trait loci analysis in a white ×red grain bread-wheat cross,Theor. Appl. Genet., 104:39–47 doi:10.1007/s001220200004
Hajjar R., and Hodgkin T., 2007, The use of wild relatives in crop improvement: A survey of developments over the last 20 years, Euphytica, 156: 1-13 doi:10.1007/s10681-007-9363-0
Himi E., Mares D.J., Yanagisawa A., and Noda K., 2002, Effect of grain colour gene (R) on grain dormancy and sensitivity of the embryo to abscisic acid (ABA) in wheat, Journal Experimental Botany, 53: 1569-1574 doi:10.1093/jxb/erf005
Hollenhorst M.M., and Joppa L.R., 1983, Chromosomal location of genes for resistance to greenbug in ‘Largo’ and ‘Amigo’ wheats, Crop Science, 23: 91-93 doi:10.2135/cropsci1983.0011183X002300010026x
Imtiaz M., Ogbonnaya F.C., Oman J., and Ginkel van M., 2008, Characterization of quantitative trait loci controlling genetic variation for preharvest sprouting in synthetic backcross-derived wheat lines, Genetics, 178(3): 1725-1736 doi:10.1534/genetics.107.084939
Kema G.H.J., Lange W., and van Silfhout C.H., 1995, Differential suppression of stripe rust in synthetic wheat hexaploid from Triticum turgidum subsp. dicoccodides and Aegilops squarrosa, Phytopathology, 85: 425-429 doi:10.1094/Phyto-85-425
Kosambi D.D., 1944, The estimation of map distances from recombination values, Ann. Eugen., 12: 172-175
Kuraparthy V., Sood S., and Gill B.S., 2008, Targeted genomic mapping of a red seed color gene (R-A1) in wheat. Crop Science, 48, 37-48 doi:10.2135/cropsci2007.08.0488tpg
Lage J., Skovmand B., and Andersen S.B., 2003, Expression and suppression of resistance to greenbug (Homoptera: Aphididae) in synthetic hexaploid wheats derived from Triticum dicoccum×Aegilops tauschii crosses, Journal of Economic Entomology, 96: 202-206 doi:10.1603/0022-0493-96.1.202
Lage J., and Trethowan R.M., 2008, CIMMYT’s use of synthetic hexaploid wheat in breeding for adaptation to rainfed environments globally, Australian Journal of Agricultural Research, 59: 461-469 doi:10.1071/AR07223
Lincoln S., Daly M., and Lander E., 1992, Constructing genetic maps with Mapmaker/EXP3.0, Whitehead Institute Techn Rep, 3rd edition, Whitehead Institute, Cambridge
Liu R.H., and Meng J.L., 2003, MapDraw: a Microsoft Excel macro for drawing genetic linkage maps based on given genetic linkage data, Hereditas (Beijing), 25: 317-321
Limin A.E., and Fowler D.B., 1993, Inheritance of cold hardiness in Triticum aestivum×synthetic hexaploid wheat crosses, Plant Breeding, 110: 103-108 doi:10.1111/j.1439-0523.1993.tb01220.x
Ginkel van M., and Ogbonnaya F., 2007, Novel genetic diversity from synthetic wheats in breeding cultivars for changing production conditions, Field Crops Research, 104: 86-94 doi:10.1016/j.fcr.2007.02.005
Ma H., Singh R.P., and Mujeeb-Kazi A., 1995, Resistance to stripe rust in Triticum turgidum, T. tauschii, and their synthetic hexaploids, Euphytica, 82: 117-124 doi:10.1007/BF00027057
McIntosh R.A., Hart G.E., Devos K.M., Gale M.D., and Rogers W.J., 1998, Catalogue of gene symbols for wheat. In: Proceedings of the ninth international wheat genetics Symposium, Vol. 5, University of Saskatchewan Extension Press, Canada
Mujeeb-Kazi A., Rosas V., and Roldan S., 1996, Conservation of the genetic variation of Triticum tauschii (Coss.) Schmalh (Aegilops squarrosa auct. non. L) in synthetic hexaploid wheats (T. turgidum L. s. lat.×T.tauschii; 2n=6x =42, AABBDD) and its potential utilization for wheat improvement, Genetic Resources and Crop Evolution, 43: 129-134 doi:10.1007/BF00126756
Nalam V.J., Vales M.I., Watson C.J.W., Kianian S.F., and Riera-Lizarazu O., 2006, Map-based analysis of genes affecting the brittle rachis character in tetraploid wheat (Triticum turgidum L.), Theor. Appl. Genet., 112: 373-381 doi:10.1007/s00122-005-0140-y
Peña R.J., Zarco-Hernandez J., and Mujeeb-Kazi A., 1995, Glutenin subunit compositions and bread-making quality characteristics of synthetic hexaploid wheats derived from Triticum turgidum×Triticum tauschii (Coss.) Schmal crosses, Journal of Cereal Science, 21: 15-23 doi:10.1016/S0733-5210(95)80004-2
Pestsova E., Ganal M.W., and Röder M.S., 2000, Isolation and mapping of microsatellite markers specific for the D genome of bread wheat, Genome, 43: 689-697 doi:10.1139/gen-43-4-689
Röder M.S., Korzun V., Wendehake K., Plaschke J., Tixier M.H., Leroy P., and Ganal M.W., 1998, A microsatellite map of wheat, Genetics, 149: 2007-2023
Sharp P.G., Kreis M., Shewry P.R., and Gale M.D., 1988, Location of bamylase sequence in wheat and its relatives, Theor. Appl. Genet., 75: 286-290 doi:10.1007/BF00303966
Sherman J.D., Souza E., See D., and Talbert L.E., 2008, Microsatellite Markers for Kernel Color Genes in Wheat, Crop Science, 48: 1419-1424 doi:10.2135/cropsci2007.10.0561
Song Q.J.,Shi J.R.,Singh S.,FickusE.W.,Costa J.M.,Lewis J.,Gill B.S.,Ward R.,and Cregan P.B., 2005, Development and mapping of microsatellite (SSR) markers in wheat, Theor. Appl. Genet., 110: 550-560 doi:10.1007/s00122-004-1871-x
Thompson J.P., Brennan P.S., Clewett T.G., Sheedy J.G., and Seymour N.P., 1999, Progress in breeding wheat for tolerance and resistance to root-lesion nematode (Pratylenchus thornei), Austr. Plant Pathology, 28: 45-52
Trethowan R.M., Villareal R., and Mujeeb-Kazi A., 1998, Pre-harvest Sprouting tolerance among synthetic hexaploid wheats, Proc. of the 8th International Symposium on Pre-harvest sprouting in cereals, Detmold, Germany
Tyler J.M., and Hatchett J.H., 1983, Temperature influence on expression of resistance to Hessian fly (Diptera: Cecidomyiidae) in wheat derived from Triticum tauschii, Journal Economic Entomology, 76: 323-326
Villareal R.L., Mujeeb-Kazi A., Rajaram S., and Toro E.d., 1994a, Morphological variability in some synthetic hexaploid wheats derived from Triticum turgidum×T. tauschii, Journal of Genetics and Breeding, 48: 7-16
Villareal R.L., Mujeeb-Kazi A., Toro E.d., Crossa J., and Rajaram S., 1994b, Agronomic variability in selected Triticum turgidum×T. tauschii synthetic hexaploid wheats, Journal of Agronomy and Crop Science, 173: 307-317 doi:10.1111/j.1439-037X.1994.tb00578.x
Villareal R.L., Sayre K., Banuelos O., and Mujeeb-Kazi A, 2001, Registration of four synthetic hexaploid wheat (Triticum turgidum/Aegilops tauschii) germplasm lines tolerant to waterlogging, Crop Science 41: 274 doi:10.2135/cropsci2001.411274x
Warner R.L., Kudrna D.A. Spaeth S.C. and Jones S.S., 2000, Dormancy in wheat- grain mutants of Chinese Spring wheat (Triticum aestivum L.), Seed Science Research, 10: 51-60
William M.D.H., Peña R.J., and Mujeeb-Kazi A, 1993, Seed protein and isozyme variations in Triticum tauschii (Aegilops squarrosa). Theor. Appl. Genet., 87: 257-263 doi:10.1007/BF00223774
Yang W.Y., Yen C., Yang J.L., and Zheng Y.L., 1999, Inheritance of resistance to Chinese wheat stripe rust races CYR30 and CYR31 in synthetic hexaploid wheat Decoy1/Aegilops tauschii 510, Southwest China Journal of Agricutural Sciences, 12 (2): 38-41 (in Chinese with English abstract)
Zhang Y., Yang W.Y., Hu X.R., Yu Y., Zou Y.C., and Li Q.M., 2004, Analysis of agronomic characters of new wheat variety Chuanmai 42 derived from synthetics (Triticum durum × Aegilops tauschii), Southwest China Journal of Agricultural Sciences, 17(2): 141-145 (in Chinese with English abstract)
. PDF(670KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Jun Li
. Huiting Wei
. Xiaorong Hu
. Baorong Lu
. Wuyun Yang
Related articles
. Synthetic hexaploid wheat
. Red-grain color
. Aegilops tauschii
. Chuanmai 42
. SSR
Tools
. Email to a friend
. Post a comment


