2.Tai’an Subcenter of National Wheat Improvement Center, Agronomy College of Shandong Agricultural University, Tai'an, 271018, P.R. China
 Author
Author  Correspondence author
Correspondence author
Molecular Plant Breeding, 2010, Vol. 1, No. 4 doi: 10.5376/mpb.2010.01.0004
Received: 06 May, 2010 Accepted: 10 Jul., 2010 Published: 28 Sep., 2010
Qi et al., 2010, Molecular tagging wheat powdery mildew resistance gene Pm21 by EST-SSR and STS markers, Molecular Plant Breeding Vol.1 No.4 (doi: 10.5376/mpb.2010.01.0004)
In order to enrich the molecular markers linked to wheat powdery mildew resistance gene Pm21 and facilitate the molecular marker-assisted selection in multi-background breeding procedure, preferred small groups (PSG) analysis was conducted using F2 segregating population cross between the wheat germplasm line CB033 with Pm21 and a common wheat variety Huixianhong (susceptible to wheat powdery mildew). One hundred and two markers revealed polymorphism between the parents when screening with 258 SSR or STS markers located on wheat homoeologous group VI, and two EST-SSR markers (Xcfe164 and Xedm129) and one STS marker (Xcinau188) showed good consistency between the genotype and the phenotype to mildew in PSG. Linkage relationship between Pm21 and molecular markers of Xcfe164, Xedm129 and Xcinau188 were then established, with the distance 3.75 cM, 1.26 cM and 0.98 cM respectively. Authenticity verification of the linkage between Pm21 and molecular markers were tested via F2: 3 families and results showed that the three markers were reliable new markers linked to Pm21. The results above would facilitate molecular marker-assisted selection in breeding program.
Background
Wheat powdery mildew caused by the obligate biotrophic fungus Erysiphe (Blumeria) graminis f.sp triticale, is one of the most serious wheat diseases, which lead to grain weight reduced, also play an negative influence on grain quality. With the promotion of semi-dwarf wheat varieties and the increasing levels of Nitrogen fertilization, the powdery mildew was more popular in China, and had become a major obstacle for wheat production (Wand et al., 2001). Identification and utilization of new powdery mildew resistance genes in breeding, have proven to be an effective means to protect the wheat from powdery mildew. The gene Pm21 located on the short arm of 6VS from Haynaldia villosa, was one of the most effective resistance genes, which conferred resistance to all races of B. graminis f. sp. tritic in China (Chen et al. 1997), Europe (Huang et al., 1997), and The powdery mildew resistance gene Pm21 has been successfully transferred to common wheat, and located on the short arm of 6VS/6AL, (Qi et al., 1995). As the fact, the translocation lines showed no obvious undesirable effects on agronomic and quality traints (Chen et al., 1995, Li et al., 2007).
Up to now, the reported molecular markers for Pm21 included RAPD, AFLP, SCAR, and STS markers. Qi et al. (1996) found the RAPD markers OPH17-1400 which could be tracked the gene Pm21, then Liu et al. (1999) converted it to SCAR (sequence characterized applied region) markers SCAR-1400 and SCAR-1265; Li et al. (2005) carried out RAPD analysis to the 6D/6V Substitution Pm930640 using 120 random primers to ,and got 5 specific markers of 6VS; Cao et al. (2006) developed a codominant marker NAU/XiBao15-902 (CINAU15-902) to identify 6VS according to the powdery mildew resistance gene sequences; Chen et al. (2006) also developed to a codominant marker NAU / XiBao16-1085 (CINAU16-1085) to identify 6VS based on the wheat LRR (leucine-rich repeat) sequence; Wang et al. (2007) found two Linked markers CINAU17-1086 and CINAU18-723 located in 0.58 ~ 0.70 and 0.00 ~ 0.45 section on 6VS respectively, when checking with 11 RGA (Resistance Gene Analogy) and 17 pairs of STS primers; Wang et al. (2007) developed a RAPD primers (OPK08910) and three AFLP primers which can be used to select the Pm21 gene from Haynaldia villosa; developed to a EST- PCR marker (XCAU127) to efficient distinguish the Pm12 and Pm21. As the RAPD markers were low stability and poor reproducibility, and the AFLP markers were technical cumbersome, expensive and difficult to automate, both were limited in plant breeding. Until now, the molecular markers used in marker-assisted selection for the Pm21 gene mainly focused on SCAR1265 (Liu et al., 1999), SCAR1400 (Liu et al., 1999), STS marker CINAU15902 (Cao et al. , 2006), CINAU161650 (Chen et al., 2006), CINAU171086 (Wang et al., 2007), CINAU18723 (Wang et al., 2007) and EST-PCR marker XCAU127.
In this study, 258 SSR or STS markers located on wheat homoeologous group VI, were used to identify the linked molecular markers for the gene Pm21, and the obtained markers will enrich the selection strategies for this gene and enhance the selection accuracy in breeding.
1 Results
1.1 Identification of wheat powdery mildew resistance
The resistant parent CB033 showed immune to mildew when the susceptible control Huixianhong fully infected. For the F2 segregation population of CB033 / Huixianhong (213 individuals), the resistant plants were 153 individuals, and the susceptible plants were 60 individuals. The difference of chi-square text was not significant (χ2 = 1.401 <χ20.05, P> 0.05), the separation ratio of resistant and susceptible plants was 3:1 (Table 1).
1.2 Screening of polymorphic primers
258 SSR or STS markers located on wheat homoeologous group VI, were used to the initial screening for the parents CB033 and Huixianhong, 102 markers in the parents revealed polymorphism. Then the 102 markers were used to the preferred small groups analysis, and two EST-SSR markers Xcfe164, Xedm129 and one STS marker Xcinau188 amplified polymorphic bands in the preferred small groups. Xcfe164 could amplify a band about 123bp size in the 10 typical resistant individuals, and could not amplify the band in the 10 typical susceptible individuals (Figure 1A); Xedm129 could amplify three bands respectively about 233 bp, 243 bp and 321 bp size in the 10 typical resistant individuals, and could not amplify the three bands in the 10 typical susceptible individuals (Figure 1B); Xcinau188 could amplify a band about 335bp size in the 10 typical resistant individuals, and could not amplify the band in the 10 typical susceptible individuals (Figure 1C).
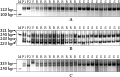 Figure 1 Amplification products by Xcfe164, Xedm129 and Xcinau188 in resistant parent CB033 (P1), susceptible parent Hui xianhong (P2), 10 typical resistant individuals and 10 typical susceptible individuals of F2 |
1.3 Linkage Analysis
To further test whether these three markers linked with the gene Pm21, F2 segregating population was used to linkage analysis. The results show that, the three markers are all closely linked with the gene Pm21, the resistant and susceptible genotypes in the F2 segregating population are consistent with the expected separation ratio (3:1) (Table 1); the linkage distance between Pm21 and molecular markers of Xcfe164, Xedm129 and Xcinau188 were 3.75 cM, 1.26 cM and 0.98 cM, respectively (Figure 2). Part F2 plants of the PCR results (Figure 3).
Table 1 Chi-square text and genetic analysis of F2 derived from the cross between CB033 and Huixianhong |
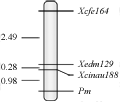 Figure 2 Linkage maps of the Pm21 in the F2 population |
Figure 3 Amplification products by Xcfe164 in resistant parent CB033 (P1), susceptible parent Hui xianhong (P2), part individuals of F2 |
1.4 Verification of molecular markers
In order to validate Xcfe164, Xcinau188 and Xedm129 were reliability, the homozygous resistant and homozygous susceptible plants which were selected from the F2 segregation population by Codominant marker NAU/XiBao15- 902(Cao et al., 2006) were planted into the F2:3 families to further resistance identification and molecular markers verification. Results show that, in the investigation of F2:3 families(130 individuals), 62 homozygous resistant plants showed immunity to powdery mildew, then 68 susceptible plants showed infection; the three markers also could amplify the corresponding marker band. So it proved that the three markers were reliable. Part F2:3 families plants of the PCR results (Figure 4A, Figure 4B).
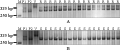 Figure 4 Amplification products by Xcinau188 in resistant parent Line CB033 (P1), susceptible parent Hui xianhong (P2), part pure resistant individuals of F2:3 |
2 Discussion
As EST were the sequence of the cloned gene fragments which were selected from the cDNA library, and STS which were based on fragments of single copy DNA sequences at both ends of specific primers and amplified specific sequences, could be directly obtained gene expression information, so the EST -SSR and STS markers were reliable. In this study, we got two EST-SSR markers (Xcfe164, Xedm129 ) and one STS markers (Xcnau188), enriched Pm21 markers system. And the genetic distances between Pm21 and molecular markers of Xcfe164, Xedm129 and Xcinau188 were all small, 3.75 cM, 1.26 cM and 0.98 cM, respectively, so they had use value in marker-assisted selection for Pm21, and can be used for marker-assisted breeding. The laboratory which used the above three markers to assisted selection of Pm21 has proven their reliability.
3 Materials and methods
3.1 Wheat materials
The wheat germplasm line CB033 (containing the powdery mildew resistance gene Pm21), the common wheat Huixianhong, CB033 and Huixianhong produced the hybrids F1, F2 and F2:3 families. The wheat germplasm line CB033 were favored by researcher Chen xiao who was from the Chinese Academy of Agricultural Sciences Department, and saved in the Tai’an Subcenter of National Wheat Improvement Center.
3.2 Primers
Selected 258 pairs of primers which all covered on wheat homoeologous group VI, included three types, genomic SSR markers, EST-SSR markers and STS markers (Yuan et al., 2010). All of the above primers were synthesized by Shanghai Biological Engineering Technology Co., Ltd.; 10 × PCR Buffer, dNTPs and Taq enzymes were Takara Biotechnology Co., Ltd. products.
3.3 Evaluation of wheat powdery mildew resistance
The susceptible parent Huixianhong was seeded into the infection line as susceptible CK , and the E15 strains were vaccinated in the infection line; when Huixianhong fully infected, surveyed the powdery mildew resistance of parents, F1, F2 and F2:3 families, and analysed the resistant and susceptible segregation of progenies.
Powdery mildew grading standards were grading standards:
0 type: Immunization, plant no lesion;
0; type: necrotic reaction, leaves have dead spots;
1 type: high resistance, small lesions (usually less than the diameter 1 mm), hyphae thin layer of green leaves can be seen, occasionally larger lesions, but through the green, sporulation little;
2 type: middle resistance, leaf lesion diameter less than 1 mm, but the mycelium layer thick, opaque green, can produce a certain amount of spores;
3 type: middle susceptible, leaf lesions more common diameter greater than 1 mm, slice thickness mycelium, sporulation capacity, but the lesion is not contiguous;
4 type: high susceptible, leaf lesion diameter greater than 1 mm, slice thickness mycelium, sporulation more contiguous lesion (see Sheng Baoqin, 1988, Plant Protection, 1: 49),
Type 0, type0;, type 1, type 2 for the resistance; type3, type4 as susceptible. The E15 strains of powdery mildew was provided by Dr. Li Hongjie who came from the Chinese Academy of Agricultural Sciences.
3.4 Extraction of total DNA
According to SDS-phenol extraction to extract the DNA of plant young leaves (Devos et al., 1993).
3.5 Detection of PCR amplification and electrophoresis
Reaction system was 15 μL, and amplification procedures used the landing PCR (touchdown PCR), then amplification save under 10 ℃(Yuan et al., 2010). Amplification equipment used by the U.S. company Bio-Rad's 9600 Thermal Cycler. PCR products were separated by 8% non-denaturing polyacrylamide gel, then stained by silver nitrate.
3.6 Markers analysis
Preferred small groups methods were used to analyze(Hao et al., 2008), extracted total DNA from the 10 typical resistant individuals (the plants of 0 or 0; type) and the 10 typical susceptible individuals (powdery mildew-level 4-type plants), then used PCR amplification and electrophoresis to search for the markers which were consistent in the phenotype and the genotype (Zong et al., 2009), further verified them in F2 segregating populations, and calculated the the linkage genetic distance between the marker and the gene (Pm21). Using chi-square test (χ2), determined the observations of the separate groups and the fitness of the expected degree. The genetic distances between Microsatellite markers and Pm21 were calculated by JoinMap software, finally, the genetic linkage map was drawn by MapDraw (Liu et al., 2003).
Author’s contributions
XLQ conducted the major part of this study including experimental design, query and synthesis of markers, analysis of experimental data, and manuscript preparation. CF participated in preliminary analysis of experimental data and manuscript preparation. YL participated in the development of the project and experimental design. DAM participated in preliminary analysis of experimental data. LJ and CGL participated in experimental preparation. HGW coordinated the project and participated in experimental design, and manuscript preparation. All authors read and approved the final manuscript.
Acknowledgements
Thank Yuanfeng Hao for his helpful comments on an earlier draft of this manuscript, Xia Wu and Liming Bao for their assistance in my experiment. And thank researcher Chen xiao for giving the wheat germplasm line CB033 and Dr. Li Hongjie provid the E15 strains of powdery mildew. All of the primers were inquired on GrainGenes2.0 (http:// wheat.pw.usda.gov/) and related References, and synthesized by Shanghai Biological Engineering Technology Co., Ltd.; 10×PCR Buffer, dNTPs and Taq enzymes were Takara Biotechnology Co., Ltd. products. This study was supported in part by the Chinese Academy of Agricultural Sciences Department. This paper is contribution from the Shandong Agricultural University and supported by the National Natural Science Foundation of china (30771349).
References
Cao A.Z., Wang X.E., Chen Y.P., Zou X.W., and Chen P.D., 2006, A sequence- specific PCR marker linked with Pm21 distinguishes chromosome 6AS, 6BS, 6DS of Triticum aestivum and 6VS of Haynaldia villosa, Plant Breed, 125: 201-205 doi:10.1111/j.1439-0523.2006.01222.x
Chen P.D., Qi L.L., Zhou B., Zhang S.Z., and Liu D.J., 1995, Development and molecular cytogenetic analysis of wheat-Haynaldia villosa 6VS/6AI. translocation lines Specifying resistance to powdery mildew, Theor. Appl. Genet., 91: 1125-1128 doi:10.1007/BF00223930
Chen X., Shi A.N., and Shang L.M., 1997, The resistance reaction of h.villosa to powdery mildew isolates and its expression in wheat background, Acta Phytopathologica sinica, 27(1): 17-22
Chen Y.P., Wang H.Z., Cao A.Z., Wang C.M., and Chen P.D., 2006, Cloning of a resistance gene analog from wheat and development of a co-dominant PCR marker for Pm21, J. Integr. Plant Biol., 48: 715-721 doi:10.1111/j.1744-7909.2006.00257.x
Devos K.M., Millan T., and Gale M.D., 1993, Comparative RFLP maps of the homeologous group-2 chromosomes of wheat, rye and barley, Theor. Appl. Genet., 85: 784-792
Hao Y.F., Liu A.F., Wang Y.H., Feng D.S., Gao J.R., Li X.F., Liu S.B., and Wang H.G., 2008, Pm23: a new allele of Pm4 located on chromosome 2AL in wheat, Theor. Appl. Genet., 117: 1205-1212 doi:10.1007/s00122-008-0827-y
Huang X.Q., Hsam S.L.K., and Zeller F.J., 1997, Identification of powdery mildew resistance genes in common wheat (Triticum aestivum L. em Thell.). IX. Cultivars, land races and breeding lines grown in China, PlantBreed, 116: 233-238 doi:10.1111/j.1439-0523.1997.tb00988.x
Li G.P., Chen P.D., Zhang S.Z., Wang X., He Z.H., Zhang Y., Zhao H., Huang H.Y., and Zhou X.C., 2007, Effects of the 6VS/6AL translocation on agronomic traits and dough properties of wheat, Euphytica, 155: 305- 313 doi:10.1007/s10681-006-9332-z
Li H., Chen X., Shi A.N., Kong F.J., Leath S., Murphy J.P., and Jia X., 2005, Characterization of RAPD markers and RFLP marker linked to powdery mildew resistant gene derived from different H.villosa, Scientia Agricultura Sinica, 38(3): 439-445
Liu R.H., and Meng J.L., 2003, MapDraw: A microsoft excel Macro for drawing genetic linkage maps based on given genetic linkage data, Hereditas, 25(3): 317-321
Liu Z., Sun Q., Ni Z., and Yang T., 1999, Development of SCAR markers linked to the Pm21 gene conferring resistance to powdery mildew in common wheat, Plant Breeding, 118: 215-219 doi:10.1046/j.1439-0523.1999.118003215.x
Qi L.L., Cao M.S., Chen P.D., Li W.L., and Liu D.J., 1996, Identification, mapping and application of polymorphic DNA associated with resistance gene Pm21 of wheat, Genome, 39: 191-197 doi:10.1139/g96-025
Qi L.L., Chen P.D., Liu D.J., Zhou B., and Zhang S.Z., 1995, The Gene Pm21—A new source for resistance to wheat powdery mildew, Acta Agronomica Sinica, (3): 57-262
Song W., Xie C.J., Du J.K., Xie H., Liu Q., Ni Z.F., Yang T.M., Sun Q.X., and Liu Z.Y., 2009, A “one-marker-for-two-genes ”approach for efficient molecular discrimination of Pm12 and Pm21 conferring resistance to powdery mildew in wheat, Mol. Breeding, 23: 357 - 363 doi:10.1007/s11032-008-9235-x
Wang C.M., Bie T.D., Chen Q.Z., Cao A.Z., and Chen P.D., 2007, Development and application of molecular markers specific to chromosome 6VS of Haynaldia villosa, Acta Agronomica Sinica, 33(10): 1595-1600
Wang X.Y., Chen P.D., and Zhang S.Z., 2001, Pyramiding and marker-assisted selection for powdery mildew resistance genes in common wheat, Acta Genetica Sinica, 28(7): 640-646
Wang Z.Y., Zhao H.M., Hong J.X., Chen L.Y., Zhu J., Li G., Peng Y.K., Xie C.J., Liu Z.Y., Sun Q.X., and Yang Z.M., 2007, Identification and analysis of four novel molecular markers linked to powdery mildew resistance gene Pm21 in 6VS chromosome short arm of Haynaldia villosa, Acta Agronomica Sinica, 33(4): 605-611
Yuan Y.Y., Wang Q.Z., Cui F., Zhang J. T., Du B., and Wang H.G., 2010, Specific loci in genome of wheat milestone parent bima 4 and their transmission in derivatives, Acta Agronomica Sinica, 36(1): 9-l6 doi:10.1016/S1875-2780(09)60027-4
Zong H., Cui F., Bao Y.G., Zhao C.H., Wang Y.H., Du B., Wang Q.Z., and Wang H.G., 2009, Developing molecular markers for the Rht gene in dwarfing germplasm line shannong 495, Journal of Triticeae Crops, 29(3): 385-389
. PDF(529KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Xiaolei Qi
. Fa Cui
. Yu Li
. Anming Ding
. Jun Li
. Guiling Chen
. Honggang Wang
Related articles
. Pm21
. Preferred small group (PSG)
. Molecular tagging
. EST-SSR
. STS
Tools
. Email to a friend
. Post a comment


