School of Sciences, Hebei University of Technology,Tianjin 300401, P.R.China

Author

Correspondence author
Molecular Plant Breeding, 2012, Vol. 3, No. 13 doi: 10.5376/mpb.2012.03.0013
Received: 18 Sep., 2012 Accepted: 20 Sep., 2012 Published: 31 Oct., 2012
The dry seeds of maize inbred line Yong 19 were radiated by 7Li heavy ion with the energy 43MeV and the doses were treated with 10 Gy, 20 Gy, 30 Gy, 40 Gy, and 50 Gy, respectively. The field data for plant height and ear position of maize inbred line induced by heavy ion from M1 (mutation generation 1) to M3 were assessed. The results showed that the plant height of the offsprings derived from maize inbred line Yong 19 had significant variation by comparing with that of the control (CK). Genetic fingerprints of M4 progenies were analyzed by using random amplified polymorphic DNAs (RAPD) method, the results revealed that there were different effects on maize genome DNA by different doses. The doses of 10 Gy and 40 Gy could yield up higher optimal selective coefficiencies that were about 0.221 4 and 0.166 4, respectively. Compared with the Control, the genetic similarity (GS) coefficients of the doses in 10 Gy and 40 Gy were less than that of other doses. The maize radiated of 10 Gy and 40 Gy were classified into two different groups, and those of 20 Gy, 30 Gy, 50 Gy were congregated into another group.
In 1927, Dr. Muller found the mutagenic effect of the rays on Drosophila and thus created new the field of radiation biology. The results of Space Flight experiments (Peterson et al., 1977) and high-altitude hot air balloon experiments (Yang and Tobias, 1979; Mei et al., 1994) demonstrated that cosmic rays had mutagenic effects on both plants and animals. Inspired by the aerospace breeding, Chinese scientist, Mr. Zengliang Yu, who applied ion beams injection techniques to crop variety improvement, has opened new multidisciplinary fields of life sciences and achieved some excellent results (Yu, 2006). As a new mutagenic source, heavy ion has some advantages compared with X-ray, including wide mutation spectra, fast stability, and small injury (Eric and Amato, 2006). At present, mutation breeding of heavy ion have been applied to the variety improvements in rice, wheat, tomato, etc. (Smith, 1967; Brunner, 1995).
With the development and application of plant molecular biology techniques, molecular marker technology has produced far-reaching impact on crop genetics and breeding (Stephen and Rita, 2008). Williams et al (1990) invented the random amplified polymorphic DNA (RAPD) technology, which can detect the DNA polymorphism is based on the polymerase chain reaction and it uses random primers to amplify genomic DNA and can detect the gene polymorphism. RAPD is a simple, fast, and effective method in the analysis of genome differences. RAPD had been applied to species classification, variety identification, molecular marker development as well as gene mapping in maize (Asif et al., 2006, Barakat et al., 2008).
In this research, the RAPD provides a basis for the understanding of radiation mutagenesis mechanism in maize inbred lines radiated by heavy ion and their descendants on the molecular level.
1 Results
1.1 The field traits from M1 to M3
The rate of seeding emergence, plant height and position of corn ear in different mutation generations the generally, the mutagenic phenotypic effects in M1 generation were less obvious but more abundant in M2 and M3 generations. Definitely it was considered important to analyze the field traits in M2 and M3 generations. The numbers of seeding emergence were counted in seven days after sowing maize inbred lines in the field. The rates of seeding emergence in M1 generation were different with the different doses of heavy ion radiation (Figure 1).
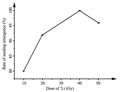
Figure 1 The rates of the seedling emergence in M1 generation
|
The plant height and position of corn ear in M2 and M3 generations were shown in table 1. The plant height of the induced lines with the doses of 10 Gy and 40 Gy were higher than that of the CK; whereas the plant height the induced lines with the doses of 20 Gy, 30 Gy and 50 Gy were shorter than that of the CK. There were exact trends in position of ear setting towards the plant height. It was clear that the trend of the trait phenotypes in M3 generation was consistent with that in M2 generation. All these data demonstrated that mutagenic effects of maize inbred line induced by heavy ion trended towards genetic stable in the M3 generation.
.png)
Table 1 Plant height and Position of ear for M2 and M3 generations |
1.2 Variance analysis on the effects of the inducing doses in M3 generation
The mutagenic effects caused by different dose were resulted from the heavy ion irradiation or experimental error with the means of variance analysis. The results showed that the variance in the doses of 10Gy and 40Gy were homogeneity of the data; the variance in the doses of 20 Gy, 30 Gy, and 50 Gy were homogeneity as well; However, two sets of the data exhibited heterogeneous.
Table 2 was the results of variance analysis among the doses of 0 Gy (CK), 10 Gy and 40 Gy. There were significant different with the comparison of each other; similarly in table 3, there were significant differences among the doses of 0 (CK) 20 Gy, 30 Gy, and 40 Gy, whereas there were not significant differences among the doses of 20 Gy, 30 Gy, and 40 Gy.
.png)
Table 2 Variance analysis of CK, 10 Gy, 40 Gy |
.png)
Table 3 Variance analysis of CK, 20 Gy, 30 Gy, 50 Gy |
The results of variance in plant height showed that the induced maize inbred lines by heavy ion 7Li were significant mutants compared with the CK .The plant height of the lines induced with the doses of 20 Gy, 30 Gy, and 50 Gy were little difference, while there was the greatest difference in plant height of the line induced by the dose of 40 Gy, following the dose of 10 Gy.
1.3 RAPD analysis of the genomic DNA of the induced maize lines
The genomic DNA of the induced maize lines were extracted by using CTAB method, and DNA integrity was detected by the electrophoresis in 0.8% agarose gel DNA concentration and purity were measured by UV1800 ultraviolet spectrophotometer. The values of OD260/OD280 were in the range from 1.8 to 2.0 (Table 4), indicating that the extracted DNA be perfect for PCR using.
.png)
Table 4 The UV detection of maize genome DNA |
27 primers were screened from 140 random primers, which could generate reproducible and clear bands. The sequences of the selected primers were shown in table 5. The 287 bands in the sizes from 250 bp to 3 000 bp were amplified by using 27 selected primers, and 89 of 287 bands exhibited polymorphic bands, accounting for 31.01% of polymorphism. The banding patterns generated by Primer 6 and primer 21 in agarose gel electrophoresis were shown in figure 2, and the results showed that the primer 6 generated two extra bands of 1 800 bp and 540 bp in length and missed a band of 410 bp in lengthin the line induced with the dose of 10 Gy. Similarly, the primer 6 enerated an extra 540 bp band in the line induced with the dose of 40 Gy. Whereas the primer 21 missed bands of 1 900 bp and 400 bp in the line induced with the dose of 10 Gy, two bands at 1 900 bp, 660 bp in the line induced with the dose of 40 Gy as well as one band of 1 900 bp in the line induced with the dose of 50 Gy.
.png)
Table 5 The 27 primers’ sequences and their amplified results |
|
.png)
Figure 2 The banding patterns generated by primer 6 and primer 21
|
1.4 Differential analysis of RAPD fingerprints and dose effect of radiation
1.4.1 The optimal selectivity coefficient and the genetic distance
The optimal selectivity coefficient M was calculated in the following function, M=nd /N*100%, Where nd is the number of differential bands between the given dose group and the control (CK); N represents total number of bands, here it was 248. The Nei’s genetic distance was calculated within the dose and CK. The optimal selectivity coefficient and the genetic distance of each dose group were listed in table 6. The results showed that the optimal selectivity coefficient of the doses of 10 Gy and 40 Gy were higher than that of the remaining doses. The genetic distance in the doses of 10 Gy and 40 Gy were 0.2214 and 0.1664, respectively.
.png)
Table 6 The optimal selectivity coefficient and the heredity distance of different dose irradiation of heavy ion 7Li
|
1.4.2 Genetic similarity
The Jaccard's Coefficients of similarity for each dose were calculated based on the RAPD data as shown in table 7. Similarities between the lines ranged from 0.794 (the lines tested in the doses of 10 Gy and 40 Gy) to 0.969 (20 Gy and 30Gy).
.png)
Table 7 The heredity similarity coefficient
|
1.4.3 Phylogenesis analysis
The dendrogram was constructed based on RAPD data by means of UPGMA (Unweighted Pair-Group Arithmetic Average Clustering) (Figure 3), and the phylogeny results presented the differences among the different doses, which was divided into three groups,the first group included the lines induced with the doses of 20 Gy, 30 Gy, 50 Gy and control (CK), the second group was the line of 40 Gy radiation , the third group was the most divergent, which was the line of the 10Gy radiation.
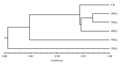
Figure 3 Dendrogram constructed based on RAPD data by UPGMA
|
2 Discussions
Luo et al studied the effects of 7Li and 12C on genetic substances of maize inbred lines (Luo et al., 2004), given variation rates in ranges from 7.7% to17.9% in M1 generation icreased with the dose increase, and RAPD analysis had proven the twin seedling derived from the irradiated progeny was a mutant type. Chen et al (2001) studied the genomic variation of seedling by nitrogen injection into alfalfa seed, and the results showed that the mutation rate ranged from 3.8% to 18.9% increased with the increase of implantation dose. In this research we got the variation rate from 4.64% to 22.14% in range, exhibited wider variation spectrum than that of the above studies, one of the reasons might be due to employing M1 generation samples in the mentioned above researches.
Although RAPD technique has been considered the poor reproducibility, it is still a useful way applying to those materials are not clear of genomic background. Our results showed that the RAPD can effectively detect genomic DNA variation, particularly identify the deletion or insertion events of genomic DNA fragments as the results of physical mutagenesis.
The present results also demonstrated that the heavy-ion radiation mutation had advantages with high rate of mutation, wide spectra, fast and stable, and low genome damage. The technique of the heavy ion mutagenesis might be an optional to build breeding populations in breeding program.
3 Materials and methods
3.1 Materials
The seeds of maize inbred line named Yong 19 were provided by Crop Research Institute, based Yongnian Countyof Hebei Province in China. The untreated seeds were set as the control (CK).
3.2 Irradiation treatment
Heavy ion irradiation experiments were carried out at Tandem Accelerator Nuclear Physics, State Key Laboratory of China Institute of Atomic Energy. The physical parameters of heavy ion 7Li were as following: Energy 43 MeV and LET 60.4 (keV/μm); the irradiation doses were set as 10 Gy, 20 Gy, 30 Gy, 40 Gy and 50 Gy
3.3 Field trials
The experimental field with sandy soil located in Yongnian County, Hebei Province in China. The plot was seeded in the width of 66 cm and the length of 5 m, each pit planted one maize seed. In order to maintain the purity of the offspring of the maize inbred line, manual bagging and forced self- pollination method were implemented in this research. The data were collected including plant height, position of ear, etc.
3.4 Genomic DNA extraction
The leaves of M3 generation lines were sampled as experimental material to extract maize genomic DNA by using CTAB method (Pal et al., 1995).
3.5 RAPD-PCR amplification
RAPD PCR reaction were done in the volume of 25 μL eppendrof tube that included 1× Buffer, 3.0 mmol/L MgCl2, 0.2 mmol/L dNTP, 0.6mmol/L Primers (Purchased from Beijing CyberSyn, China), 80 ng template, 1 U Tap enzymes, and ddH2O adding to final volume.
PCR amplification procedure followed pre-denaturization at 94℃ for 10 min, and then 35 cycles of denature- zation at 94℃ for 1 min, annealing at 37℃ for 30 sec., extending at 72℃ for 90 sec., finally extending at 72℃ for 10 min. PCR amplification was performed in the PCR instrument model TC-3000 (Techne, Jenway). RAPD amplification products were separated and scored in 1.5% agarose gel electrophoresis.
3.6 RAPD data analysis
Polymorphic bands were calculated by POPGEN32 software, and Nei's genetic distance (Nei and Li, 1979) was adopted to assess the relationship between radiation dose and the percentage of polymorphic bands (PPB). Genetic similarity coefficient (GS) was calculated by the NTSYSpc2.1. Dendrogram was constructed by UPGMA clustering analysis method based on GS data.
Author’s contributions
JPG and DFL carried out the molecular work and drafted the manuscript. TGC,JL and XZW participated in data collection and organized experimental materials. DFL and CLH participated in the design of the field trials and performed the statistical analysis. YZ, JPG and YFC conceived this study revised the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This work was supported by a grant from Science and Technology Support Program of Hebei Province (09220102D)
References
Asif M., Rahman M., Zafar Y., 2006, Genotyping analysis of six maize (Zea may L.) hybrid using DNA fingerprinting technology, Pakistan Jounral of Botany, 38 (5): 1425-1430
Barakat M.N., Milad S.I., EI-Sgafei A.M., Khatab S.A., 2008, Genetic analysis and identification of RAPD markers link to northern corn leaf blight disease resistance in a white maize population, Jounral of King
Abdulaziz University - Meteorology, Environment and Arid Land Agriculture Sciences, 20(1): 45-61
Brunner H., 1995, Radiation induced mutations for plant selection, Applied Radiation and Isotopes, 46(.6/7): 589-594
Eric J. H., and Amato J. G., 2006, Radiobiology for the radiologist, 6th edition, Lippincott Williams & Wilkins, Philadelphia, USA, pp.5-11
Maliga P., Klessig D.F., Cashemore A.R., Gruissem W., Varner J.E., 1995, Methods in plant molecular biology: A laboratory course manual, Cold Spring Harbor Lab Press, USA, pp.167-190
Mei M., Qiu Y., He Y., Bucker H., Yang C.H., 1994, Mutational Effects of Space Flight on Zeal Mays Seeds, Advances in Space Research, 14(10): 33-39
Nei M., and Li W.H., 1979, Mathematical model for studying genetic variation in terms of restriction endonucleases, Proc. Natl. Acad. Sci., 76(10): 5269-5273
Peterson D. D., Benton E.V., Tran M., Yang T., Freeling M., Craise L., Tobias C.A., 1977, Biological effects of high-LET particles on corn-seed embryos in the Apollo- Soyuz Test Project--Biostack III experiment, Life Sciences and Space Research, 15: 151-155
Smith H.H., 1967, Reletive biological effectiveness of different types of ionizing radiations: Cytogenetic effects in maize, Radiation Resesrch Supplement, 7: 190-195
Stephen P.M., and Rita H.M., 2008, Molecular plant breeding as the foundation for 21st century crop improvement, Plant Physiology, 147(3): 969-977
Williams J.G., Kubelik A. R., Livak K.J., Rafalski J.A., Tingey S.V., 1990, DNA polymorphisms amplified by arbitrary primers are useful as genetic markers, Nucleic Acids Research, 18(22): 6531-6535
Yang T.C, and Tobias C.A., 1979, Potential use of heavy ion radiation in crop improvement, Gamma Field Symposia, 18: 141-151
Yu Zengliang, 2006, Introduction to ion beam biotechnology, Springer, New York and Berlin, pp.193-196
 Author
Author  Correspondence author
Correspondence author

.png)
.png)



