The genetic linkage map is the important basis for QTL analysis of plant traits and gene mapping and cloning. Choosing the appropriate genetic markers are considered as the key in the process of the construction of genetic maps. However, the amount of traditional morphological genetic markers is very limited, and it is easy to be influenced by environment changing as well as difficult to use directly. Yet molecular markers have significant advantages overcoming shortcomings mentioned above and have been widely used in the genetic linkage map of cabbage (Song et al., 1991; Ajisaka et al., 1995; Nozaki, et al., 1997; Matsumoto et al., 1998; Zhang et al., 2000; Kim et al., 2006).
AFLP, as a molecular marker, combined the advantages of the RFLP and the RAPD, has a plenty of polymorphism, in formativeness and reproducibility. So that AFLP was considered as one of an idea tools to construct high-density molecular genetic map in cabbage (Wang, 2004; Zhang et al., 2005; Sun et al., 2006; Zhang et al., 2004; Geng et al., 2007).
cDNA-AFLP derived from the AFLP technology is kind of RNA fingerprinting approach, which mole- cular polymorphism was generated by the expression of gene coding region. Nowadays it became a simple and practical new method to construct transcriptome map (Brugmans et al., 2002; Pan et al., 2007; Fan et al., 2008).
Caixin (Brassica rapa var. Parachinensis) is a special landrace of the flowering cabbage in southern China. The edible parts are the flower stalk so that the important traits of bolting and initial flowering stage are very closely associated with the yield and commodity. Although there are many cabbage genetic linkage maps to be constructed in the past years, cDNA-AFLP based linkage map was not yet to be reported in Caixin as a relative of cabbage family. In this study a pair of extremely late or early flowering Caixin varieties were employed to make a cross to generate an F2 population that included 92 individual plants applied to construct a cDNA-AFLP based genetic linkage map, the research purposes aimed to provide reliable genetic information for mapping and cloning of the flowering gene as well as comparative genomic research.
1 Results and Analysis
1.1 cDNA-AFLP analysis
Fifteen primer combinations were selected for cDNA-AFLP amplification (Figure 1), which gene- rated 229 bands in total with a range from 8 to 21 and average of 7.5 bands each pair (Table 1). 113 polymorphic fragments were scored with the average bands from 4 to 14 per pair in range, of which the 34 bands (32%) came from parents. 7 (6.5%) bands from the maternal parent, 8 (7.5%) from paternal parent, whereas the remaining 64 (54%) bands were not associated with any parent. The size of the bands was distributed in the range from 100 bp to 500 bp in length. The Chi-square (χ2) tests indicated that the 104 loci were not in line with the expected Mendelian segregation ratio (1:2:1), the segregation distortion was up to 45.4%.

Figure 1 The selective amplification in F2 population by E2M1 primer combination
|
|
.png)
Table 1 cDNA -AFLP analysis in the F2 population
|
1.2 The cDNA-AFLP based linkage map constructed
Four linkage groups were generated with the LOD score 3.0 that contained 67 loci , which span 334 cM of the genome (Table 2) The linkage group (LG)1 contained 33 markers spanning 190 cM; while the LG2 even distributed 27 markers spanning 72.4 cM with mean distance 2.68 overall and the maximum distance was 6.80 cM between markers. The LG3 span 38.2 cM with 5 markers and LG4 only included 2 markers (Figure 2). We thought the cDNA-AFLP marker trended to a clustering distribution in the constructed linkage map that need to adopt other types of molecular marker to overcome this problem possible caused by the separation distortion. The further studies are in progress.
.png)
Table 2 The characteristics of genetic linkage map of Caixin
|
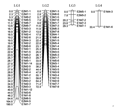
Figure 2 The cDNA-AFLP map constructed with 67 loci in F2 population of 92 F2 individuals of Caixin
|
Although AFLP and cDNA-AFLP markers both can be used for constructing cabbage molecular genetic map at the level of genome and transcriptome, AFLP would be much more popular used in practices because of its excellent advantages (Vos et al., 1995; Yu et al., 2003; Wang et al., 2004; Zhang et al, 2005; Sun et al., 2006; Wang et al., 2009; W et al., 2011). Due to DNA molecular markers being not a gene, lots of markers used to link the agronomic traits to construct a precise linkage map. In fact, it was not easy making the AFLP markers associated with the phenotype, which might be one of the reasons why many genetic maps in cabbage were not yet to be in practices. Another reason might be insufficient AFLP markers distributed on non-coding centromeric regions of chromosome (Vuylsteke et al., 1999; zhang et al., 2005).
There were some examples of cDNA-AFLP markers used in construction of transcriptome map in Arabidopsis, potato, cabbage, and cotton and so on (Brugmans et al., 2002; Trindade et al., 2004; Fan et al., 2008; Pan et al., 2007). The advantages of cDNA-AFLP transcriptional markers were clear that the polymorphism was generated from functional genomic sequence (Brugmans et al., 2002; Trindade et al., 2004).
The segregation distortion is a common genetic phenomenon and do not have solid relationshipwith the marker types, although some studies showed that AFLP markers had the highest proportion of segregation distortion (Ding et al., 2012). In this study, 104 of 229 markers being segregation distortion were found accounting 45.4%the lower than that of reported by Zhang et al (2005), Zhang et al (2005) and Zhang et al (2008), while higher than that of others such as Wang et al (2004), Geng et al (2007), San et al (2009), Wang et al (2009) as well as the cDNA-AFLP results of cabbage (Fan et al., 2008).
It has been known that segregation distortion should be related to some factors, such as gametogenesis, homologous recombination, chromosome behavior, transposons and so on. It was not yet to know whether the significant segregation distortion was connected to the extremely different flowering parent or the smaller population size or developmental abnormalities, which might be further investigated.
In view of the genetic map constructed based on varieties hybridization that may display the complete genetic information about the target traits, this would be one of the reason in this study that an F2 population was developed in order to maintain the whole separated diversity. Due to the limited individuals of the population and limited molecular markers used in this research, the results in this study could not generated linkage groups in coincidence with cabbage chromosome number (2n=10 ).
3 Materials and Methods
3.1 F2 population developed in this research
The mapping population derived from a cross between extremely early flowering cultivar, GUIZAO and extremely late flowering cultivar, GUIWAN, as parents to generate the F1 seeds and then self- pollinated to develop F2 population that contained 92 individuals. The parents and F2 populations were simultaneously planted and transplanted in the field at the three-leaf stage. The initial flowering date was recorded as days of bolting while picked young leaves to be stored in liquid nitrogen for RNA extraction.
3.2 cDNA-AFLP analysis
TRIzol kit (the product of the Life Technologies Company) was used to extract RNA from parents and F2 individual leaves. SMARTTM PCR cDNA Synthesis Kit (the product of the Clontech Company) was used to reverse transcript RNA to cDNA. cDNA-AFLP connector and primers were synthesized by Sangon Biotech (Shanghai) Co., Ltd and their sequences showed in Table 1. Chemicals such as EDTA, DEPC Acrylamide Bis-adrylamide, urea, ammonium persulfate, and TEMED were also purchased from Sangon Biotech Company. Taq DNA polymerase was purchased from the Invitrogen Biotechnology Co., Ltd. AFLP protocol was followed up the report used in our lab before (Wei et al., 2011).
3.3 Data analysis
ABH method was employed for data processing. AFLP maker is a dominant marker, so we assigned 'A' to homozygote representing the allele from maternal parent, 'B' to homozygote representing the allele from paternal parent, while 'H' to heterozygote that carried both alleles from parents and '-' representing missing data for the individuals at this locus. The name of cDNA-AFLP markers was tagged in accordance with the size of fragments following the name of primers combination.
3.4 Linkage group construction
Linkage analysis and map construction was done by Joinmap4.0 software with Kosambi's mapping function.
Author's contributions
SWD fulfilled major part of this research including varieties collection, experimental design and cDNA-AFLP work, data analysis and manuscript preparation. ZJH and YCF accomplished cDNA-AFLP experimant. LWJ participated in trait phenotyping and LHL assisted some Lab and field work in this research.
Acknowledgements
This project was jointly supported by Guangxi Science Foundation (2010GXNSFA013084) and Agricultural Scientific Institutions of Guangxi Basic Research Project (2012JZ06 and 2012YT05).
References
Ajisaka H., Kuginuki Y., Hida K., Enomoto S., and Hirai M., 1995, A linkage map of DNA markers in Brassica campestris, Breeding Science, 45(Suppl.): 195
Ding G.D., Yang M., Li X.M., Shi L., and Xu F.S., 2012, Segregation distortion analysis of molecular markers in RIL population of Brassica Napus, Journal of Agricultural Science and Technology, 14 (2): 56-61
Fan S.Y., Le J.G., Cheng G.J., and Wu C.J., 2008, Construction of Chinese cabbage-pak-choi transcriptome map with cDNA-AFLP techniques, Scientia Agricultura Sinica, 41(6): 1735-1741
Geng J.F., Hou X.L., Zhang X.W., Cheng Y., Li Y., Yuan Y.X., Jiang W.S., Han Y.P., and Yao Q.J., 2007, Construction of a genetic linkage map in non-heading Chinese cabbage (Brassica campestris ssp. chinensis) using a doubled haploid (DH) population, Journal of Nanjing Agricultural University, 30(2): 23-28
Kim J.S., Chung T.Y., King G.J., Jin M., Yang T.J., Jin Y.M., Kim H.I., and Park B.S., 2006, A sequence-tagged linkage map of Brassica rapa, Genetics, 174: 29-39
http://dx.doi.org/10.1534/genetics.106.060152
Lou P., Zhao J.J., Kim J.S., Shen S .X., Carpio D.P.D., Song X.F., Jin M., Vreugdenhil D., Wang X.W., Koornneef M., and Bonnema G., 2007, Quantitative trait loci for flowering time and morphological traits in multiple populations of Brassica rapa, Journal of Experimental Botany, 58: 4005-4016
http://dx.doi.org/10.1093/jxb/erm255
Matsumoto E., Yasui C., Ohi M., and Tsukada M., Linkage analysis of RFLP markers for clubroot resistance and pigmentation in Chinese cabbage (Brassica rapa ssp. pekinensis), Euphytica, 1998, 104(2): 79-86
http://dx.doi.org/10.1023/A:1018370418201
Nozaki T., Kurnazak A., Koha T., Ishikawa K., and Ikehashi H.,1997, Linkage analysis among loci for RAPDs, isozymes and some agronomic traits in Brassica compestris L., Euphytica, 95: 115-123
http://dx.doi.org/10.1023/A:1002981208509
Shan X.Z., Li Y., Gao S.Y., Ma C.Y., Liu T.K., Zhu Y.Y., and Hou X.L., 2009, Molecular genetic map of Brassica Campestris ssp.Chinensis, Acta Botanica Boreali-Occidentalia Sinica, 29(6): 1116-1121
Shi et al., 2011, Genetic Diversity of 30 Cai-xins (Brassica rapa var. parachinensis) Evaluated Based on AFLP Molecular Data, Molecular Plant Breeding Vol.2 No.7 (doi: 0.5376/mpb.2011.02.0007)
Song K.M., Suzuki J.Y., Slocum M.K., Williams P.H., and Osborn T.C., 1991, A linkage map of Brassica rapa (syn. campestris) based on restriction fragment length polymorphism loci, Theoretical and Applied Genetics, 82: 296-304
http://dx.doi.org/10.1007/BF02190615
Sun X.F., Cben Z.D., and Li D.Q., 2006, Construction of AFLP based genetic linkage map of Chinese cabbage (Brassica campestris L.ssp. pekinensis) in F2 population derived from combination between resistant and susceptive to Tipburn, Fenzi Zhiwu Yuzbong (Molecular Plant Breeding), 4(1): 65-70
Trindade L.M., Horvath B.M., van Berloo R., and Visser R.G.F., 2004, Analysis of genes differentially expressed during potato tuber life cycle and isolation of their promoter regions, Plant Science, 166(2):423-433
http://dx.doi.org/10.1016/j.plantsci.2003.10.016
Vos P., Hogers R., Bleeker M., Reijans M., van de Lee T., Hornes M., Frijters A., Pot J., Peleman J., and Kuiper M., 1995, AFLP: a new technique for DNA fingerprinting, Nucleic Acid Research, 21: 4407-4414
http://dx.doi.org/10.1093/nar/23.21.4407
Vuylsteke M., Mank R., Antonise R., Bastiaans E., Senior M.L., Stuber C.M., Melchinger A.E., Luebberstedt T., Xia X.C., Stam P., and Kuiper M., 1999, Two high-density AFLP® -linkage maps of Zea mays L.: analysis of distribution of AFLP markers, Theor. Appl. Genet., 99: 921-935
http://dx.doi.org/10.1007/s001220051399
W. Xinhua, L.Yang, C. Huoying., 2011, A linkage map of pak-choi (Brassica rapa ssp. chinensis) based on AFLP and SSR markers and identification of AFLP markers for resistance to TuMV,Plant Breeding, 130(2): 275-277
http://dx.doi.org/10.1111/j.1439-0523.2010.01811.x
Wang M., Wang J., Li H.B., Lin R.P., and Piao Z.Y., 2009, Constructed an AFLP genetic linkage map of Chinese cabbage with resistant and susceptible F2 lines, Acta Agriculturae Boreali-Sinice, 24(2): 64-70
Wang M., Zhang F.L., Meng X.D., Liu X.Y., and Fan Z.C., 2004, A linkage map construction for Chinese cabbage based on AFLP markers using DH population, Acta Agriculturae Boreali-Sinica,19(1): 28-33
Yu S.C., Wang Y.J., and Zheng X.Y., 2003, Construction and analysis of a molecular genetic map of Chinese cabbage, Scientia Agricultura Sinica, 36: 190-195
Zhang C.G., and Gan L., 2005, Using small page to optimize cDNA-AFLP, Journal of Huazhong Agricultural University, 24 (2): 113-116
Zhang F.L., Wang M., Liu X.C.,Zhao X. Y., and Yang J. P., Quantitative trait loci analysis for resistance against Turnip mosaic virus based on a doubled haploid population in Chinese cabbage, Plant Breed, 127(1): 82-86
Zhang L.G., Wang M., Chen H., and Liu L., 2000, Construction of RAPDs molecular genetic map of Chinese cabbage, Acta Botanica Sinica, 42 (5): 484-489
Zhang W.F., Yu S.C., Yu Y.J., Zhang F.L., Zhang D.S., Zhao X.Y., and Xu J.B., 2007, Cloning and sequence analysis of fragments related to chilling requirement late bolting in Chinese cabbage, Molecular Plant Breeding, 5(4): 559-564
Zhang X.F., Wang X.W., Lou P., Zhang X.W., Wang Y.Q., Yuan Y.X., Zhao J.J., and Sun R.F., 2005, An AFLP-based genetic linkage map of Chinese cabbage using double haploid (DH) population,Acta Horticulturae Sinica, 32(3): 443-448
Zou Y. M., Yu S.C., Zhang F.L., Yu Y.J., Zhao X.Y., and Zhang D.S., 2009, cDNA-AFLP analysis on transcripts associated with bolting in Brassica rapa L. ssp. Pekinensis, Hereditas, 31(7): 755-762

 Author
Author  Correspondence author
Correspondence author

.png)


