1 Introduction
Leguminosae family contains species of Plants, Herbs, Shrubs, and Trees. Legumes are used as crops, forages and green manures; they also synthesize a wide range of natural products such as flavours, drugs, poisons and dyes. The legume family is the third largest family of angiosperms (
Mabberley, 1997) with approximately 730 genera and over 19,400 species worldwide (
Lewis et al., in press). Legumes are able to convert atmospheric nitrogen into nitrogenous compounds useful to plants which is achieved by the presence of root nodules containing bacteria of the genus Rhizobium. These bacteria have a symbiotic relationship with Legumes, fixing free nitrogen for the plants; in return legumes supply the bacteria with a source of fixed carbon produced by photosynthesis. This enables many legumes to survive and compete effectively in poor nitrogen conditions. Legumes are noticeably absent to poorly represent in mesic temperate habitats, including many arctic and alpine regions and the understory of cool temperate forests. The predilection of legumes for semi-arid to arid habitats is related to a nitrogen-demanding metabolism, which is thought to be an adaptation to climatically variable or unpredictable habitats whereby leaves can be produced economically and opportunistically (
McKey, 1994;
Wojciechowski et al., 2004). Leguminosae family is further classified into three subfamilies; Fabaceae (Papilionaceae), Caesalpiniaceae and Mimosaceae.
1.1. matK gene
The
matK gene, formerly known as
orfK, is emerging as yet another gene with potential contributions to plant molecular systematics and evolution (
Johnson and Soltis, 1994, 1995;
Steele and Vilgalys, 1994;
Liang and Hilu, 1996). The gene, ~1500 base pairs (bp), is located within the intron of the chloroplast gene
trnK, on the large single-copy section adjacent to the inverted
repeat. Further, the molecular information generated from matK hasbeen used to resolve phylogenetic relationships from shallow to deep taxonomic levels (Johnson and Soltis, 1994; Hayashi and Kawano, 2000; Hilu et al., 2003; Cameron, 2005). In addition to the importance of matK in plant phylogenetics, it is also the only putative group II intron maturase encoded in the chloroplast genome (Neuhaus and Link, 1987). Maturases are enzymes that catalyze the removal of nonautocatalytic intron from premature RNAs. Maturases generally contain three domains: a reverse-transcriptase (RT) domain, domain X (the proposed functional domain), and a zinc-finger-like domain (Mohr et al., 1993). The 3′ region of matK has homology to the domain X of mitochondrial group II intron maturases (Neuhaus and Link, 1987). This region of matK also lacks indels (Hilu and Liang, 1997), indicating evolutionary constraint and conservation of function. Among higher plants, matK is the only plastid gene containing this putative maturase domain (Neuhaus and Link, 1987). The matK gene stands out among plastid genes used in plant systematics in its distinct mode and tempo of evolution. The rate of substitution in matK is three times higher at the nucleotide level and is six times higher at the amino acid level than that of rbcL (Johnson and Soltis, 1994; Olmstead and Palmer, 1994), denoting it as a fast or rapidly evolving gene (Soltis and Soltis, 2004). The accelerated rate of amino acid substitution in matK is due to almost even distribution of substitution rates among the three codon positions compared with most protein-coding genes where the rates are skewed toward the third codon position.
1.2. NCBI (The National Center for Biotechnology Information)
NCBI is part of the United States National Library of Medicine (NLM), a branch of the National Institutes of Health. The NCBI houses a series of databases relevant to biotechnology and biomedicine. Major databases include GenBank for DNA sequences, Protein, Genome, EST etc. All these databases are available online through the Entrez search engine(
http://www.ncbi.nlm.nih.gov/).
1.3. DNA (Deoxyribonucleic acid) / Nucleotide
DNA is a molecule that encodes the genetic instructions used in the development and functioning of all known living organisms and many viruses. Along with RNA and proteins, DNA is one of the three major macromolecules essential for all known forms of life. DNA is well-suited for biological information storage, since the DNA backbone is resistant to cleavage and the double-stranded structure provides the molecule with a built-in duplicate of the encoded information (
http://www.ncbi.nlm.nih.gov/nuccore/).
1.4. Protein
Proteins are large biological molecules consisting of one or more chains of amino acids. Proteins perform a vast array of functions within living organisms, including catalyzing metabolic reactions, replicating DNA, responding to stimuli, and transporting molecules from one location to another (
http://www.ncbi.nlm.nih.gov/protein/).
2 Materials and Methods
In this paper we have considered around 266 species which are found in Gujarat state of India (Sagar Patel et al., 2013). Further we searched each species in NCBI database and finally found around 148 species’ information like DNA, Protein and other useful information of leguminosae family (Sagar Patel et al., 2014). Further we have only considered matK gene sequences of DNA and Protein sequences. Evolutionary analysis done in MEGA 5 software with two different methods one with Maximum Likelihood (ML) method with Bootstrap method and other method is Neighbor-Joining (NJ) method(Figure 2) and Table 1 described detailed information of each species which is considered for this study with accession number of NCBI database.
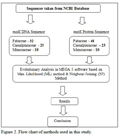
Figure 2 Flow chart of methods used in this study
|
.gif)
Table 1 Species details from NCBI database
|
3 Results
A. Resultsof DNA MATK Gene Sequeces
Caesalpiniaceae Subfamily
As shown in Figure 3 which is the result of Max.Likelihood method and Figure 3.1, which is the result of NJ method, species of Cassia, Caesalpinia, Delonix and Bauhinia genus are related as per morphological characters or botanical classifications. Even single species of same genus is not distantly related; they are all related very closely. This is very strange and rare result that two different tree method share same result even it don’t change any sequential order also. So, there might be strong sequence similarity between them so that they remain unchanged in two different results which are done with two different methods.
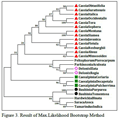
Figure 3 Result of Max.Likelihood Bootstrap Method
|
Mimosaceae Subfamily
As shown in Figure 4 which is the result of Max.Likelihood method and Figure 4.1, which is the result of NJ method, species of Albizia and Acacia genusare related as per morphological characters or botanical classifications. Even single species of same genus is not distantly related; they are all related very closely. This is very strange and rare result that two different tree method share same result even it don’t change any sequential order also. So, there might be strong sequence similarity between them so that they remain unchanged in two different results which are done with two different methods.
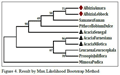
Figure 4 Result by Max.Likelihood Bootstrap Method
|
Fabaceae Subfamily
As shown in
figures 5 and
5.1, species of genus
Medicago, Trigonella, Sesbania, Crotalaria, Butea and few species of
Vigna genus are related as per morphological characters or botanical classifications. In
figure 5,
Clitoria ternatea found in the
Vigna genus, so that it may be homologus species to other species of
Vigna genus. There might be strong sequence similarity of
Clitoria ternatea with other species of
Vigna.
While in figure 5.1 species like Vigna radiata and Vigna unguiculata share common node but they are found distantly with their other species of same genus. Vigna aconitifolia doesn’t share common node with any species of Vigna species. While there is no change if we compare two figures 5 and 5.1 and we observed that there are many species of same genus which have not changed in both results and their sequential order remains unchanged. So, there might be strong sequence similarity between species that they remain unchanged in two different results which are done with two different methods.

Figure 5 Result by Max.Likelihood Bootstrap Method
|
B. Results of PROTEIN MATK Gene Sequences
Caesalpiniaceae Subfamily
In Figure 6, species of Cassia, Caesalpinia, Delonix and Bauhinia genus are related as per morphological characters or botanical classifications but Parkinsonia aculeata found between species of Delonix genus, so it may be closely related with species of Delonix genus.
In Figure 6.1, it shows that even single species of Cassia, Caesalpinia, Delonix and Bauhinia genus are not distantly related; they are all related very closely and their sequential order also remain unchanged when we compare these two results which follow different methods of analysis.
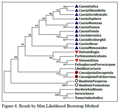
Figure 6 Result by Max.Likelihood Bootstrap Method
|
So, the NJ method gives the more accurate result of evolutionary relationship of Caesalpiniaceae subfamily as per morphological or taxonomical classification where the results were based on matK protein sequences and species remain unchanged and fall within their genus which is true as per their taxonomical classification.
Mimosaceae Subfamily
As shown in Figure 7, species of genus Albizia and species of Acacia genus are related as permorphological characters or botanical classifications except Albizia lebbeck which is not closely related with Albizia,whereas species of
Albizia genus are correctly related with each other as per botanical classification.
As shown in Figure 7.1, Albizia and Acacia genus are related as per morphological characters or botanical classifications except Acacia senegal and but Pithecellobium dulce fallwithin Albizia genus which is homologous with other species of Albizia. If we compare Figures 4 and 4.1, it was observed that species like Acacia farnesiana and Acacia nilotica; their sequential order remains unchanged in both results. So,there might be strong sequence similarity between them, and the results remain unchanged in two different methods.

Figure 7 Result by Max.Likelihood Bootstrap Method
|
Fabaceae Family
As shown in above
Figures 8 and 8.1, species of
Medicago, Sesbania, Butea, Lathyrus, Vicia, Canavalia, Crotalaria and few species of
Vigna and
Trigonella are related as per morphological characters or botanical classifications except
Tephrosia purpurea and
Vigna unguiculata which are distantly related to other species of their respective genera. Most of genera shown accurate result of their species compared with their botanical classification and evolutionary results which is discussed above and match correctly with each other.

Figure 8 Result by Max.Likelihood Bootstrap Method
|
The results of bootstrap by Maximum Likelihood andNJ methods showed striking similarity in the phylogenetic relationship among the species. This is an indication of similar sequence similarity among the species.
4 Conclusion
Reconstructing the phylogenetic relationships of the Leguminosae is essential for understanding the origin and diversification of this ecologically and economically important family of angiosperms. Strong phylogenetic signal from matK has rendered it an invaluable gene in plant systematics and evolutionary studies at various evolutionary depths. Species belonging to Leguminosae family which is further divided into three subfamilies like Fabaceae (Papilionaceae), Mimosaceae and Caesalpiniaceae; This study shows that based on morphological characters species of each subfamily are classified differently and they fall in different groups based on DNA and Protein sequences of matK. In this study we observed that as per the botanical classification species are classified differently based on their morphological features like species’ flower color, size and shape, types and arrangements of Stipules, size of plant etc. But this study focus on evolutionary relationship of species in the Leguminosae Family species based on DNA and Protein sequences of matK gene with Multiple sequence alignment by Maximum likelihood and Neighbor Joining methods where we observed that some species belonging to same genus are fall verynearly as per botanical classification which is correct as per both botanical and evolutionary relationship but we observed that few species are distantly related even if they are from same genus. Even some species’ position also remains unchanged in both Maximum likelihood and Neighbor Joining methods.
Acknowledgement
We are heartily thankful to Prof. (Dr.) P.V. Virparia, Director,GDCST, Sardar Patel University, Vallabh Vidyanagar, forproviding us facilities for the research work.
Cameron K. M.. 2005. Leave it to the leaves: a molecular phylogenetic study of Malaxideae (Orchidaceae). American Journal of Botany 92: 1025-1032.
Ems S. C. Morden C. W. Dixon C. K. Wolfe K. H. dePamphilis C. W. Palmer J. D.. 1995. Transcription, splicing and editing of plastid RNAs in the nonphotosynthetic plant Epifagus virginiana. Plant Molecular Biology 29: 721-733.
Gadek, P. A., P. G. Wilson, AND C. J. Quinn. In pressPhylogenetic reconstruction in Myrtaceae using matK, with particular reference to the position of Psiloxylon and Heteropyxis. Australian Systematic Botany.
Harborne, J.B. 1994. Phytochemistry of the Leguminosae. In Phytochemical Dictionary of the Leguminosae, eds Bisby,F.A. et al. London: Chapman & Hall
Hayashi K. Kawano S. 2000. Molecular systematics of Lilium and allied genera (Liliaceae): phylogenetic relationships among Lilium and related genera based on the rbcL and matK gene sequence data. Plant Species Biology 15: 73-93.
Hilu K. W. Borsch T. Müller K. Soltis D. E. Soltis P. S. Savolainen V. Chase M. W. Powell M. P. Alice L. A. Evans R. Sauquet H. Neinhuis C. Slotta T. A. B. Jens G. R. Campbell C. S. Chatrou L. W. 2003. Angiosperm phylogeny based on matK sequence information. American Journal of Botany 90: 1758-1776.
Hilu KW, Liang H: The matK gene: sequence variation and application in plant systematics. American Journal of Botany 1997, 84:830-839.
Johnson L. A. Soltis D. E. 1994. matK DNA sequences and phylogenetic reconstruction in Saxifragaceae s. str. Systematic Botany 19: 143-156.
Martin F. Wojciechowski,matt Lavin,michael J. Sanderson. A Phylogeny Of Legumes (Leguminosae) Based On Analysis Of The Plastid Matk Gene Resolves Many Well-supported Subclades Within The Family.
Michelle M. Barthet, Hilu KW : Expression of matK: functional and evolutionary implications. American Journal of Botany 2007, vol. 94 no. 8 1402-1412.
Mohr G. Perlman P. S. Lambowitz A. M. 1993. Evolutionary relationships among group II intron-encoded proteins and identification of a conserved domain that may be related to maturase function. Nucleic Acids Research 21: 4991-4997.
Neuhaus H. Link G. 1987. The chloroplast tRNALys (UUU) gene from mustard (Sinapsis alba) contains a class II intron potentially coding for a maturase-related polypeptide. Current Genetics 11: 251-257.
Patel SS, Vaidya MB, Shah DB (2014) Homology Modelling of Conserved rbcL Amino Acid Sequences in Leguminosae Family. J Data Mining Genomics Proteomics 5: 154. doi:10.4172/2153-0602.1000154.
Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 5 (Tamura, Peterson, Stecher, Nei, and Kumar 2011).
Polhill, R.M. & Raven, P.H. (eds) 1981. Advances in Legume Systematics. Royal Botanic Gardens, Kew
Sagar Patel and Shah, 2014, Phylogeny in Few Species of Leguminosae Family Based on matK Sequence, Computational Molecular Biology, Vol.4, No.6 1-5 (doi:10.5376/cmb.2014.04.0006).
Sagar Patel, and Hetalkumar Panchal. 2014, Bioinformatics Information of Leguminosae Family in Gujarat State, International Journal of Agriculture, Environment & Biotechnology: Vol 7, Issue 1, Pages:11-15.
Sagar Patel, Hetalkumar Panchal. Evolutionary studies of few species belonging to Leguminosae family based on RBCL gene. Discovery, Volume 9, Number 22, January 2014, Pages: 38-50. ISSN 2278 – 5469, EISSN 2278 – 5450.
Sagar Patel, Panchal H., 2013. Leguminobase: A Tool To Get Information Of Some Leguminosae Family Members From NCBI Database in Journal of Advanced Bioinformatics Applications and Research: Vol 4, Issue 3, 2013, Pages. 54-59. ISSN 0976-2604. Online ISSN 2278 – 6007.
Sagar Patel, Panchal H., Smart J., Anjaria K., 2013. Distribution of Leguminosae family members in Gujarat State of India: Bioinformatics Approach in International Journal of Computer Science and Management Research, Pages- 2184-2189 Vol 2 Issue 4 April 2013, ISSN 2278-733X.
Sagar Patel, Panchal H., Smart J., Anjaria K., 2013. Species Information Retrieval Tool: A Bioinformatics tool for Leguminosae family in International Journal of Bioinformatics and Biological Science: Vol.1 n.2 Pages.187-194 June, 2013 Print ISSN 2319-5169.
Smartt, J. & Simmonds, N.W. (eds) 1995. Evolution of Crop Plants. Harlow: Longman Scientific & Technical
Soltis D. E. Soltis P. S.. 2004. Amborella not a “basal angiosperm”? Not so fast. American Journal of Botany 91: 997-1001.
Steele, K. P., AND R. Vilgalys. 1994. Phylogenetic analyses of Polemoniaceae using nucleotide sequences of the Plastid gene matK. Systematic Botany 19:126-142.
Sugita M. Shinozaki K. Sugiura M.. 1985. Tobacco chloroplast tRNALys (UUU) gene contains a 2.5-kilobase-pair intron: an open reading frame and a conserved boundary sequence in the intron. Proceedings of the National Academy of Sciences, USA 82: 3557-3561.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, and Kumar S (2011) MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology and Evolution (submitted).
 Author
Author  Correspondence author
Correspondence author
.gif)

.gif)

.gif)












