Analysis of Insertion Copy Number and Integration Site of T-DNA in the Genome of Transgenic High Oelic Rapeseed (Brassica napus L.) 
 Author
Author  Correspondence author
Correspondence author
Plant Gene and Trait, 2011, Vol. 2, No. 3 doi: 10.5376/pgt.2011.02.0003
Received: 09 Nov., 2011 Accepted: 10 Dec., 2011 Published: 17 Jan., 2012
Chen et al., 2011, Analysis of Insertion Copy Number and Integration Site of T-DNA in the Genome of Transgenic High Oelic Rapeseed (Brassica napus L.), Plant Gene and Trait, Vol.2, No.3 15-22 (doi: 10.5376/pgt.2011.02.0003)
In order to acquire the information about the insertion copy number and integration site of T-DNA in the genome of transgenic high oleic acid rapeseed (Brassica napus L.), we isolated the genomic DNA from the transgenic line W-4, the T2 generation plants, then the transgenic genomic DNA was digested with BamHI prior to conducting Southern bolt with the probe of a segment of NPT II labeled with Dig. The results showed that only one copy of T-DNA was detected to be integrated into the genome of transgenic line W-4. To isolate the flanking sequence of both right border and left border of T-DNA insertion in the genome, the thermal asymmetric interlaced PCR (TAIL-PCR) was employed by using three or four nested specific primers designed based on the sequence of the vector pCNFIRnos and a short arbitrary degenerate primer (AD1 or AD2) , respectively. The PCR products corresponding to flanking sequences of the right and left border were specifically amplified. The flanking sequence of right border is 470 bp in length which includes a 290 bp genomic sequence and a 180 bp vector sequence based on analysis of VecScreen, while the left border flanking sequence is 641 bp in length including a 365 bp genomic sequence and a 276 bp vector sequence. Further sequence alignment analysis revealed that the 180 bp sequence is identical to the RB border of pCNFIRnos vector, which exists a 62 bp deletion, whereas the later of 276 bp is better identical to the left border of pCNFIRnos vector besides a change from G to A happened. Therefore, it was suggested that the integration of the T-DNA in the genome of transgenic line W-4 should be a kind of vector backbone-free integration. Furthermore, there is no any information about homologous sequence of acquired flanking sequences found in the genome based on the Blastn analysis, which implied that the T-DNA should be integrated into non-coding region of the genome. In conclusion, we considered that the information about the T-DNA insertion copy number, integration site and flanking sequences in the genome of the transgenic rapeseed W-4 might be very useful for the biosafety assessment of genetically modified rapeseed and for the indentification of the transgenic high oleic acid rapeseed.
Rapeseed, belonging to Brassica genus of Cruciferae family, is one of the world's four major oil crops as well as an important model crops for genetic transformation. Past two decades at least 50 genes were transformed into Brassica napus rapeseed (Brassica napus L.), the transferred genes were mainly those traits related to insect resistance (Du et al., 2007; Lin et al., 2002; Wang et al., 2005), disease resistance (Lan et al., 2000; Ma et al., 2008), herbicide resistance (Graef et al., 2007; Peng et al., 1998; Sakhno et al., 2008), male sterility (He et al., 2003; Engelke et al., 2010), fatty acid synthesis and quality improvement (Knutzon et al., 1999; Stoutjesdijk et al., 2000; Mietkiewska et al., 2008; Shi et al., 2001) and stress tolerance (Wang, 2006). More than 10 GM canola varieties in Canada and other countries had been approved to plant in the commercial production (Lu, 2006, cited from the Chinese journal of Zhongguo Youliao Zuowu Xuebao, 27(4): 106-110). Figure 1 The band signals of the Southern blotting in this study
GM rapeseed needs to be done strictly for a series of GMO safety assessment before GM rapeseed is approved to release in commercial production. One of principal contents is to analyze the molecular characteristics of genetically modified plants in detail, including the T-DNA insertion copy number, T-DNA integration mode, information about the insertion sites and their flanking sequences, in order to facilitate the biological safety evaluation (Wu et al., 2010). In view of random and un-reproducibility of T-DNA integration sites in the process of transformation, the flanking sequence information of the T-DNA integration sites can be used for detecting specificity of genetically modified events, which would be facilitate to monitor and manage the GMO products in commercialization. In addition, exogenous gene copy number in transgenic plants is one of the factors to affect the targeted gene expression and genetic stability, which would be the important criteria to determine whether or not the GM plants have potential values in practical application.
Jiangsu Academy of Agricultural Sciences has successfully introduced the reverse repeat sequence expression cassette of oleic acid desaturase gene (fad2) into the B. napus varieties by Agrobacterium-mediated transformation, obtaining genetically modified high oleic acid germplasm, called as W-4 (Chen et al., 2009). At present T3 generation seeds have been harvested, which showed that the lines had stable and high oleic acid traits by analysis of fatty acid composition and offspring derived from sexual crossing presented a dominant inheritance (data to be published). However, it is not yet to be clear about T-DNA copy number and integration sites and other relevant information in the genome of transgenic rapeseed. In order to provide the prerequisite information for GMO biosafety assessment of transgenic high-oleic rapeseed, in this study we attempted to analyze the T-DNA insertion copy number by Southern blotting genomic DNA of transgenic high-oleic rapeseed using NPTâ…¡ gene fragment labeled with DIG as probe, as well as to analyze the T-DNA integration modes, sites and their flanking sequences by using TAIL-PCR amplification with the degenerate primer and nested sequence-specific primer combinations designed based on the sequence of pCNFIRnos vector.
1 Results and Analysis
1.1 T-DNA copy number analysis
The genomic DNAs of four individuals of W-4 transgenic rapeseed that were PCR-positive as well as DNA of non-transformed plants (negative control) were performed by Southern blotting with DIG-labeled fragment of the NPTâ…¡ gene as probes, respectively. The results showed that hybridization signals were detected in the genomic DNA of four PCR-positive plants, a distinctive hybridization band. Since these four plant lines were derived from the transformed W-4, the positions of hybridization bands were completely the same line. Whereas the negative control was not detected any hybridization signal (Figure 1). This result demonstrated that the target gene has been integrated into the rapeseed genome. As the vector pCNFIRnos used for transformation has only a BamHâ… restriction site in the T-DNA region (Chen et al., 2006), we predicted that genetically modified high oleic acid rapeseed W-4 should be integrated single copy T-DNA based on the number of band in Southern hybridization.
![]()
1.2 TAIL-PCR amplification products
The right border flanking sequences of T-DNA insertion sites of high oleic acid transgenic lines W-4 were amplified by four round TAIL-PCR amplification with four pairs of primers in turn following RBF1/AD1, RBF2/AD1, RBF3/AD1 and RBF4/AD1. A distinct bright band of about 500 bp in size was amplified in the fourth round PCR amplification comparing PCR products of the second, third and fourth rounds amplification by gel electrophoresis (Figure 2 lane 1, 2, 3). The products with the intervals of 70 bp was amplified by the nested primers between RBF2 and RBF3, as well as and 130 bp interval appeared between RBF3 and RBF4. Therefore, according to the changes of the product sizes in the second, third and fourth round amplifications, it might imply that the product of 500 bp should be specific amplification products. Likewise, In the amplification of the left border flanking sequences of T-DNA insertion sites, A distinct bright band of about 700 bp in size was amplified in the third round TAIL-PCR amplification using three pairs of primers LBF1/AD2, LBF2/AD2 and LBF3/AD2, by comparing PCR products of the second, third and rounds amplification by gel electrophoresis (Figure 2 lane 4, 5), after 3 TAIL-PCR amplification performed. The products with the intervals of 80 bp was amplified by the nested primers between LBF2 and LBF3, according to the changes of the product sizes in the second, third round amplifications, it might predict that the product of 700 bp should be specific amplification products.
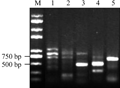 Figure 2 TAIL-PCR products by gel electrophoresis |
1.3 Sequence analysis of right border PCR products
The 500 bp band of the right border in the fourth round PCR amplification was recovered, cloned and sequenced, we obtained a 470 bp sequence by artificially removing the sequence of vector pEASY-T1. Blast analysis showed that the sequence from 1st to 180th base was the vector's sequences (Figure 3) as well as from the 181st-470th base belonged to genomic sequence of rapeseed based on VecScreen online analysis (http://www.ncbi.nlm.nih.gov/VecScreen/VecScreen.html). Comparing the obtained sequence with the T-DNA right border sequence of pCNFIRnos was found that there was highly homologous between 180 bp of vector sequence and the T-DNA right border sequence. However, the 62 bases from the 181st to 242nd base that including the right border was lost in the genome of the transgenic rapeseed (Figure 4).
Figure 3 Result of right border flanking sequence by VecSreen |
Figure 4 Alignment between the sequence of the right border integrated into genome of transgenic line and the sequence of the right border of vector pCNFIRnos |
1.4 Sequence analysis of left border PCR products
The band of the left border in the third round PCR amplification was recovered, cloned and sequenced, we obtained a 641 bp sequence by artificially removing the sequence of vector pEASY-T1. The results showed that 276 bp from the 366th to 641st base had highly homologous with the vector sequence based on online VecScreen and the sequence from the 1st to 365th base belonged to rapeseed genome (Figure 5). Comparing the obtained 276 bp sequence with the T-DNA right border sequence of pCNFIRnos was found that the sequence had highly homologous with the T-DNA left border sequence. However, the second base site of juncture between LB border and genome of rapeseed had a base conversion from G to A (Figure 6).
Figure 5 Result of left border flanking sequence by VecScreen |
Figure 6 Alignment between the sequence of the left border integrated into genome of transgenic line and the sequence of the left border of vector pCNFIRnos |
1.5 Flanking sequence verification
Two primers were designed, LBF and RBR, according to T-DNA flanking sequences of rapeseed, which were combined with LBF1, LBF2, LBF3 and RBF1, RBF2, RBF3, RBF4. PCR amplification were conducted by using transgenic rapeseed genomic DNA as template. The results showed that the product of left border were amplified in size of 800 bp, 700 bp and 600 bp, respectively, while of right border were amplified in size of 750 bp, 650 bp, 600 bp and 450 bp, respectively, which all were fully consistent with the expecting sizes (Figure 7). It was proved that the genomic sequence of the transgenic rapeseed we obtained should be connected with both borders of vector, which was the flanking sequence of the T-DNA insertion sites.
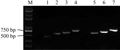 Figure 7 Validation of the flanking sequences |
2 Discussions
Currently, most of GM crops were achieved through Agrobacterium-mediated method. The process of Agrobacterium-mediated transformation is a kind of the perfect process of occurrence in nature. The specific sequence known as T-DNA can be accurately cut and integrated into the genome of recipient plant to be delivered to the offspring. As a natural plant genetic transformation system, there are some advantages including: full structure of transformed foreign DNA, stable of integration sites, low copy number and less variation of the integrated foreign gene structure, etc. (Brenda et al., 2009; Dai et al., 2001). In contrast, gene bombarding transform technology often leads to multiple-copy integration (Shou et al., 2004). Studies have shown that insertion of foreign genes with low copy number (1 or 2) tends to have a better exogenous gene expression, while integration of multi-copy number will lead to instability of gene expression and even the silencing phenomenon of transgene (Iyer et al., 2000; Vaucheret et al., 1998). Genetic stability of transgene should be a prerequisite for commercial application of genetically modified plants. Therefore, the identification of genetically modified plants not only to determine whether the foreign gene was integrated into the recipient genome, but also to analyze how much transgenic copy number was there. In practice we should choose a single copy inserted into plants as much as possible to be applied in the breeding program.
In this research, Southern blotting results revealed that single T-DNA copy was integrated into genome of genetically modified high oleic acid rapeseed W-4, in other words the oleic acid desaturase gene (fad2) of the inverted repeat sequence expression cassette (Chen, 2006) was integrated with a single copy into the W-4 genome, which effectively inhibited the functions of fatty acid desaturation enzyme in preventing the synthesis of oleic acid to linoleic acid. This is the basis of W-4 exhibiting high-oleic trait to be stable inheritance.
In this study, the flanking sequence of T-DNA insertion sites of the transgenic W-4 genome was performed. According to the sequence characteristics of left and right borders of T-DNA region in the original transformation vector pCNFIRnos, the length of the nested primers were extend more than 30 bp. And also due to the right T-DNA is the napin promoter sequence and taking into account of existence of homologous sequences in the rapeseed genome, so we added an additional round PCR amplification for amplifying the right border flanking sequences in order to obtain specific amplifying products. The results proved that the experimental design should be correct.
We had amplified 365 bp and 290 bp of the T-DNA left and right border sequences, respectively, which were validated by Tail-PCR amplification for the flanking sequences around the insertion site, the flanking sequences were proved to be the sequences of the T-DNA insertion site, which found no highly homologous sequences deposited in Brassica genome database. So, we can use these flanking primers for qualitative or quantitative testing of transgenic events. In addition, according to the morphological observations of GM rapeseed and non-transgenic receptor, there was no significant difference between them, indicating that the transgenic insertion site might be in the non-coding region.
The study also found that transgenic rapeseed W-4 had a complete T-DNA left border in genome, but only a base substitution occurring, whereas 62 bases missing including the RB border in the right border region. It has been little reported that such a large fragment was lost. Base deletion or substitution used to occur in the T-DNA border sequences and flanking sequence of the insertion site in the host genome. In most of transgenic plants, the probability of occurrence of loss in T-DNA left border would usually be greater than that of the right border (Tzfira et al., 2004). T-DNA right border avoided to be attacked by exonuclease because of the right border binding with the VirD2 proteins during integration process (Bundock and Hooykaas, 1996). Missing VirD2 protein will result in the right border missing (Michielse et al., 2004).
However, It has found that the probability of T-DNA right border missing in the transgenic tobacco was greater than that the left border (Krizkova and Hrouda, 1998; Salomon and Puchta, 1998). Lee et al (2006) found right borders of pCAMBIA-1300 T-DNA border sequences in Agrobacterium-mediated transformation of fungi easily resulted in missing the sequence of right border. In this study we adopted a binary vector pCNFIRnos that the basic skeleton is pCAMBIA-2300, comparing with pCAMBIA-1300, the only difference is that the former T-DNA using kanamycin resistance gene as selectable marker gene, while the late is the hygromycin resistance gene. So, it still needs to be verified in much more transgenic plants whether the reason could be due to the characteristics of vector sequence. In this study, the characteristics of T-DNA integration in genetically modified high oleic acid rapeseed is so-called vector backbone-free integration, which was reported that the probability of this integration in Agrobacterium-mediated transformation was only 30% that also related to the variety of the binary vector based on published literatures (Ye et al., 2008).
In summary, the results obtained in this study might provide important molecular information for assessing biological safety of transgenic high oleic rapeseed W-4 as well as for detecting the genetically modified high oleic acid rapeseed.
3 Materials and Methods
3.1 Materials used in this research
Genetically modified high oleic acid rapeseed W-4 (about 84% oleic acid content) (Brassica napus L.) developed by transforming Brassica napus variety, Westar (Canola, a kind of double low variety), mediated by Agrobacterium in this laboratory. DNA was extracted from the seedlings of the T2 generation individuals and wildtype Westar.
3.2 Enzymes and reagents
Restriction endonuclease BamHâ… and Tag enzyme (TaKaRa code R001M) are TaKaRa company's products. Cloning vector, pEASY-T1, was purchased from Quanshijin Biotechnology Co., Ltd. DIG-DNA labeling kit, Southern blot solution and DIG-detection kit are the Roche (Indianapolis, IN, USA) products. HybondTM-N+nylon membrane is Amersham Biosciences products. Upstream and downstream primers of NPTâ…¡ gene were synthesized by Beijing Sanbo Biotechnology Company, as well as other primers were synthesized by Shanghai Invitrogen Company.
3.3 Nucleic acid hybridization
Preparation of hybridization probes: The plasmid pCAMBIA2300 DNA used as template, fragment of the NPTâ…¡ gene was DIG-labeled by PCR amplification using PCR DIG Labeling Mix, which is the probe of 688 bp in size. NPTâ…¡-specific primers were shown in table 1.
Table 1 List of the primers in this study |
DNA blotting: a large number of genomic DNA of rapeseed was extracted by using CTAB method. 30 μg DNA of each sample were completely digested with BamHâ… for 12 hours. Digesting products were separated on 0.8% agarose gel electrophoresis, and then DNA gel inclusions were transferred to nylon membrane by using the capillary method.
Southern hybridization: Southern blotting was followed the product's manual. The pre-hybridization and hybridization between the probe and transferred nylon membrane were performed in 40℃, and then the membrane was been washing, detecting, exposing, developing and fixing with CDP-Star detection solution.
3.4 Flanking sequences of T-DNA insertion sites by TAIL-PCR amplification
Random primers AD1 and AD2 were designed following the method of Liu et al (1995). The nested specific primers were design based on left or right border region sequences of T-DNA of transgenic expression vector pCNFIRnos (Table 1). The nested primers were combined with the primers of AD1 and AD2 for amplification. PCR reactions were performed in total of 20 μL of reaction mixture containing: 10 ng DNA template, 2 μL buffer, 0.2 mmol/L dNTPs, 0.25 mmol/L MgCl2, 0.25 μmol/L primers, and 1 U of Tag enzyme. The primer combinations of the first round PCR were RBF1/AD1for right border primer and LBF1/AD2 for left border primer combinations. In the second round of PCR, the products from the first round PCR were diluted by 50 times and taken 1 μL as a template, primer combinations were RBF2/AD1 and LBF2AD2. In the third round of PCR, the products from the second round PCR were further diluted by 50 times and taken 1 μL as a template, primer combinations were RBF3/AD1 and LBF3AD2. In the fourth round of PCR amplification, primer combinations RBF4/AD1 for right border region were performed only. The procedure of TAIL-PCR performance was followed with Liu et al (1995). PCR products of each round PCR were detected by electrophoresis, the bands with similar expected sizes were chosen to be recovered, cloning and sequencing.
3.5 Flanking sequence verification
Specific primers LBF and RBF designed based on the flanking sequences of the gene sequence of rapeseed were used to make primer combinations with LBF1, LBF2, LBF3 and RNF1, RBF2, RBF3, RBF4, respectively, PCR amplification was performed using genomic DNA of transgenic rapeseed as template. In total of 20 μL reaction mixture containing: 10 ng DNA template, 2 μL buffer solution, 0.2 mmol/L dNTPs, 0.25 mmol/L MgCl2, 0.25 μmol/L primers, 1 U of Tag enzyme. PCR procedure was performed: 95℃ for 4 min. pre-denature, 30 cycles following 95℃ for 45 s denature, 58℃ for 45 s annealing, 72℃ for 45 s extension and the final 72℃ for 5 min extension.
Authors' contributions
Song Chen, Aijuan Shen, Xiaoying Zhou, Weihua Long and Maolong Hu are the persons who carried out this experiment. Jiefu Zhang participated in some in lab work and involved the data analysis and in field work; Song Chen wrote the manuscript and Huiming Pu revised the manuscript. Cunkou Qi conceived the project and designed the experiments as well as wrote and revised manuscript. All authors had read and agreed the final text.
Acknowledgments
This research was jointly sponsored by the 948 Project of MOA (Q04) and Supporting program of Jiangsu province (BE2009304) Authors appreciated Dr Dong of Tsinghua University for his help to finish the Southern Bloting experiments and also sincerely thank for two anonymous reviewers with their critical comments. In this paper we mentioned some chemical and reagent suppliers and sequencing service providers, that doesn't mean we would like to recommend or endorse the production of theirs.
Reference
Brenda A.L., Prakash N.S., Way M., Mann M.T., Spencer T.M., and Boddupalli R.S., 2009, Enhanced single copy integration events in corn via particle bombardment using low quantities of DNA, Transgenic Res., 18(6): 831-840
http://dx.doi.org/10.1007/s11248-009-9265-0 PMid:19381853
Bundock P., and Hooykaas P.J.J., 1996, Integration of Agrobacterium tumefaciens T-DNA in the Saccharomyces cerevisiae genome by illegitimate recombination, Proc. Natl. Acad. Sci., USA, 93(26): 15272-15275
http://dx.doi.org/10.1073/pnas.93.26.15272
Chen S., Pu H.M., Zhang J.F., Gao J.Q., Chen F., Long W.H, Hu M.L., and Qi C.K., 2009, Identification of high oleic acid germplast from the T_2 Progeny of the transgenic Brassica napus L., Jiangsu Nongye Xuebao (Jiangsu Journal of Agricultural Sciences), 25(6): 1234-1237
Chen S., Zhang J.F., Chen F., Chen X.J., Long W.H., Pu H.M., and Qi C.K., 2006, Construction of a seed-specific expression ihpRNA vector targeting the fad2 gene of Brassica napus L., Zhongguo Youliao Zuowu Xuebao (Chinese Journal of Oil Crop Sciences), 28(3): 251-256
Dai S.H., Zheng P., Marmey P., Zhang S.P., Tian W.Z., Chen S., Beachy R.N., and Fauquet C., 2001, Comparative analysis of transgenic rice plants obtained by Agrobacterium-mediated transformation and particle bombardment, Mol. Breeding, 7(1): 25-33
http://dx.doi.org/10.1023/A:1009687511633
Du W.M., Guan R.Z., Wang J.F., Zhang H.S., Tang S.Y., and Dong H.B., 2007, Studies on transformation of a serine proteinase inhibitor gene Ds TI2 from descurainia sophia into Brassica napus L., Fenzi Zhiwu Yuzhong (Molecular Plant Breeding), 5(6): 765-770
Engelke T., Hirsche J., and Roitsch T., 2010, Metabolically engineered male sterility in rapeseed (Brassica napus L.), Theor. Appl. Genet., 122(1): 163-174
http://dx.doi.org/10.1007/s00122-010-1432-4 PMid:20821307
Graef F., Stachow U., Werner A., and Schutte G., 2007, Agricultural practice changes with cultivating genetically modified herbicide-tolerant oilseed rape, Agricultural Systems, 94(2): 111-118
http://dx.doi.org/10.1016/j.agsy.2006.09.008
He Y.H., Xiong X.H., Guan C.Y., Li X., Lin L.B., Chen S.Y., Liu Z.S., Li W.B., Zhong J., Liu C.L., and Zhou X.Y., 2003, The pTA29-Barnase chimeric gene transformation of Brassica napus mediated by Agrobacterium, Zuowu Xuebao (Acta Agronomica Sinica), 29(4): 615-620
Iyer L.M., Kumpatla S.P., Chandrasekharan M.B., and Hall T.C., 2000, Transgene silencing in monocots, Plant Mol. Biol., 43(2-3): 323-346
http://dx.doi.org/10.1023/A:1006412318311 PMid:10999414
Knutzon D.S., Hayes T.R., Wyrick A., Xiong H., Davies H.M., and Voelker T.A., 1999, Lysophosphatidic acid acyltransferase from coconut endosperm mediates the insertion of laurate at the sn-2 position of triacylglycerols in lauric rapeseed oil and can increase total laurate levels, Plant Physiology, 120(3): 739-746
http://dx.doi.org/10.1104/pp.120.3.739 PMid:10398708 PMCid:59311
Krizkova L., and Hrouda M., 1998, Direct repeats of T-DNA integrated in tobacco chromosome: characterization of junction regions. Plant J., 16(6): 673-680
http://dx.doi.org/10.1046/j.1365-313x.1998.00330.x PMid:10069074
Lan H.Y., Wang C.H., Zhang L.H., Liu G.Z., Wan L.L., Chen Z.H., and Tian Y.C., 2000, Studies on transgenic oilseed rape (Brassica napus) plants transformed with β-1,3-glucanase and chitinase genes and its resistance to Sclerotinia sclerotiorium, Shengwu GongCheng Xuebao (Chinese Journal OF Biotechnology), 16(2): 142-146
PMid:10976313
Lee M.H., and Bostock R.M., 2006, Agrobacterium T-DNA-mediated integration and gene replacement in the brown rot pathogen Monilinia fructicola, Curr. Genet., 49(5): 309-322
http://dx.doi.org/10.1007/s00294-006-0059-0 PMid:16468040
Lin L.B., Guan C.Y., Zhou X.Y., He Y.H., and Yang Z.X., 2002, Stable heredity and efficient expression of Bt Insecticidal protein gene in the transgenic rapeseed and its insect-resistant activity, Zuowu Xuebao (Acta Agronomica Sinica), 28(2): 175-178
Liu Y.G., Mitsukawa N., and Oosumi T., 1995, Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR, Plant Journal, 8(3): 457-463
http://dx.doi.org/10.1046/j.1365-313X.1995.08030457.x PMid:7550382
Ma L.L., Zhai R., Gao X.W., He D., Shao M., and Wang Q., 2008, Transgenic rape with hrf2 gene encoding harpin (Xooc) resistant to Sclerotinia sclerotinorium, Zhongguo Nongye Kexue (Scientia Agricultura Sinica), 41(6): 1655-1660
Michielse C.B., Ram A.F.J., Hooykaas P.J.J., and van den Hondel C.A.M.J.J., 2004a, Agrobacterium-mediated transformation of Aspergillus awamori in the absence of full-length VirD2, VirC2, or VirE2 leads to insertion of aberrant T-DNA structures, J. Bact., 186(7): 2038-2045
http://dx.doi.org/10.1128/JB.186.7.2038-2045.2004 PMid:15028687 PMCid:374399
Mietkiewska E., Hoffman T., Brost J.M., Giblin E.M., Barton D.L., Francis T., Zhang Y., and Taylor D.C., 2008, Hairpin-RNA mediated silencing of endogenous FAD2 gene combined with heterologous expression of Crambe abyssinica FAE gene causes an increase in the level of erucic acid in transgenic Brassica carinata seeds, Mol. Breeding, 22(4): 619-627
http://dx.doi.org/10.1007/s11032-008-9204-4
Peng R.W., Zhou X.R., Wang J.L., Fang R.X., Chen Z.H., and Mang K.Q., 1998, Transgenic oilseed rape plants expressing barstar gene and bar gene, Yichuan Xuebao (Acta Genetica Sinica), 25(1): 74-79
Sakhno L.A., Gocheva E.A., Komarnitskii I.K., and Kuchuk N.V., 2008, Stable expression of the promoterless bar gene in transformed rapeseed plants, Cytology and Genetics, 42(1): 16-22
Salomon S., and Puchta H., 1998, Capture of genomic and T-DNA sequences during double-strand break repair in somatic plant cells, EMBO J., 17(20): 6086-6095
http://dx.doi.org/10.1093/emboj/17.20.6086 PMid:9774352 PMCid:1170935
Shi D.Q., Zhou Y.H., Hu Z.M., Zhang L.H., Liu G.Z, and Chen Z.H., 2001, Introduction of trans desaturase gene into Brassica napus L. via particle bombardment and obtaining of transgenic plants, Nongye Shengwu Jishu Xuebao (Journal of Agricultural Biotechnology), 9(4): 359-362
Shou H.X., Frame B.R., Whitham S.A., and Wang K., 2004, Assessment of transgenic maize events produced by particle bombardment or Agrobacterium-mediated transformation, Mol. Breeding, 13(2): 201-208
http://dx.doi.org/10.1023/B:MOLB.0000018767.64586.53
Stoutjesdijk P.A., Hurlstone C., Singh S.P., and Green A.G.., 2000, High-oleic acid australia Brassica napus and B. juncea varieties produced by co-supression of endogenous delta-12 desaturases, Biochemical Society Transactions, 28(6): 938-940
http://dx.doi.org/10.1042/BST0280938 PMid:11171263
Tzfira T., Li J., Lacroix B., and Citovsky V., 2004, Agrobacterium T-DNA integration: molecules and models, Trends Genet., 20(8): 375-383
http://dx.doi.org/10.1016/j.tig.2004.06.004 PMid:15262410
Vaucheret H., Béclin C., Elmayan T., Feuerbach F., Godon C., Morel J.B., Mourrain P., Palauqui J.C., and Vernhettes S., 1998, Transgene-induced gene silencing in plant, Plant J., 16(6): 651-659
http://dx.doi.org/10.1046/j.1365-313x.1998.00337.x PMid:10069073
Wang J., Chen Z., Du J., Sun Y., and Liang A., 2005, Novel insect resistance in Brassica napus developed by transformation of chitinase and scorpion toxin genes, Plant Cell Rep., 24(9): 549-555
http://dx.doi.org/10.1007/s00299-005-0967-3 PMid:16028062
Wang Y., Zeng Y.L., He B., Qin L., Li J.Y., Gao Y., and Zhang F.C., 2006, Research of NHX gene transformation in Brassica napus by Agrobacterium tumefaciens, Zuowu Xuebao (Acta Agronomica Sinica), 32(2): 278-282
Wu X.X., Zhang B.B., Wang Z.K., Jiang C.T., and Li W.B., 2010, Safety management and assessment of genetically modified organisms, Zuowu Zazhi (Crops), 4: 1-4
Ye X., Williams E.J., Shen J., Esser J.A., Nichols A.M., Petersen M.W., and Gilbertson L.A., 2008, Plant development inhibitory genes in binary vector backbone improve quality event efficiency in soybean transformation, Transgenic Res., 17(5): 827-838
http://dx.doi.org/10.1007/s11248-008-9169-4 PMid:18253857
. PDF(799KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Song Chen
. Jiefu Zhang
. Huiming Pu
. Aijuan Shen
. Xiaoying Zhou
. Weihua Long
. Maolong Hu
. Cunkou Qi
Related articles
. Rapeseed ( Brassica napus L.)
. Transgenic high oleic acid rapeseed
. Gene insertion copy
. Integration site
. T-DNA
Tools
. Email to a friend
. Post a comment


