 Author
Author  Correspondence author
Correspondence author
Tree Genetics and Molecular Breeding, 2012, Vol. 2, No. 2 doi: 10.5376/tgmb.2012.02.0002
Received: 20 Dec., 2011 Accepted: 17 Jan., 2012 Published: 14 Mar., 2012
Wang et al., 2012, Molecular Evolutional Characteristics of the Plant ITS Sequences in Buxus Genus, Tree Genetics and Molecular Breeding, Vol.2, No.2 8-14 (doi: 10.5376/tgmb.2012.02.0002)
There are many plant species belonging to the genus Buxus. The taxonomy in the genus is far more confusing, especially endemic to Chinese pearl boxwood (Buxus sinica var. parvifolia), a rare endangered species in China, which remains controversial in phylogenetics and classification yet. In this research, we chose five plant species in the Buxus genus and five populations of endangered Chinese pearl boxwood (Buxus sinica var. parvifolia) to analyze rDNA ITS sequences. By combining the other nine species of Buxus genus reported in the GenBank and using ITS sequence of Pachysandra terminalis as reference, we did alignment analysis for the ITS sequences of 18 species of the Buxus genus. The results indicated that nucleotide substitution in ITS sequences of Buxus plants should be far more transition occurring than transversion being. In the region of ITS-1, the numbers of bases transiting are almost similar as that of bases transversing, while in the region of ITS-2 the numbers of base transiting are more than that of base transversing and the same situation takes place in the region of 5.8S. However, the ratio of base substitutions in the regions of ITS-1 and ITS -2 or 5.8S did not reach significant difference based on the calculations following the Juks & Cantor’s One Parameter Model and Kimura’s Two Parameter Model in statistics. The values of the ratio calculated by Two Parameter Model were a little bit larger than that by One Parameter Model due to transition took place more than transversion happened in the region of ITS-2. Based on detecting the relative rate of molecular evolution of different clustering groups in Genus Buxus, the results presented that there was an obvious difference of molecular evolution rate of ITS sequences among the clusters in Buxus plants. In this study, it suggested that Chinese pearl boxwood (B. sinica var. parvifolia ) should be a sister species instead of a variant of B. sinica according to the relative rates of detection and phylogenetic analysis. Furthermore, there was close genetic relationship between B. sinica var. parvifolia and B. henryi.
The Buxus is the largest genus in the family of Buxaceae, representing approximately two-third plants in this family. The texture of wood is known as fineness, density and good looking, suitable for making sculpture, art and other crafts. It is also commonly used to be ornamental plants, which is a good tree species for garden and landscape greening due to strong resistance to cold. Some species in this family containing boxwood alkaloids has an high medicinal values for curing coronary heart disease, etc. However, phylogenetic relationships and classification of species in the genus Buxus are more confusing yet. There were different descriptions for species recorded in different flora, it is difficult to distinguish some species with similar morphologies in the practical application that would bring some trouble for scientific research and practical cultivation. Endemic to Chinese Pearl boxwood (Buxus sinica var. Parvifolia), the taxonomic status and phylogenetic relationship have yet remained controversial.
Pearl boxwood was published as a variant of B. sinica (Zheng, 1980). Lin (2004) found that Pearl boxwood had distinct differences in variety of phenotypes from B. sinica by studying specimens of Pearl boxwood. Whereas Pearl boxwood was very similar with B. rugulosa, both of them might be the same species. In addition, the quantitative classification of the genus Buxus in China carried out and comparisons with the morphological classification had done (Yan et al., 2002).
In recent years, molecular analysis is more widely applied in the phylogenetic study of Buxus genus. Wang et al (2008) analyze isozymes of peroxidase and esterase and their genetic relationship among three species in Buxus by polyacrylamide gel electrophoresis. Lv and Ji (2009) carried out fingerprint analysis of Pearl boxwood clones using isozyme and ISSR. Jiang (2008) studied the genetic diversity of two natural populations of Pearl boxwood using RAPD markers. Huang et al (2008) also conducted genetic structure and diversity of five natural populations and one cultivated population of Pearl boxwood using RAPD and ISSR markers to explore the taxonomical status of related taxa of the Pearl boxwood.
ITS, inter-transcribed spacer, is located in the middle between 18S and 26S in rDNA, which was divided into two parts namely ITS-1 and ITS-2, by the 5.8S region. ITS has a feature of conservative in length and high variability of nucleotide sequence. Therefore, there are abundant of variable sites and informative sites. ITS has become an important molecular marker systems for studying classification and evolution under the level of genus in angiosperm and has already applied in phylogenetics in a variety of plant taxa. But it is yet to be reported genus Buxus phylogenetics and taxonomic status of Pearl boxwood through the ITS sequences.
In this research, we directly sequenced rDNA ITSs of five species from Genus Buxus and five population of endangered Chinese Pear boxwood to analyze the characteristics of ITS and the rules of variation. By combining the ITS data of other species in Buxus deposited in GenBank, we analyzed ITSs of 18 species in Buxus to study the law of molecular evolution as well as to further understand the classification and phylogenetic status of Chinese Pear boxwood.
1 Results and Analysis
1.1 Nucleotide substitutions of ITS in Buxus plants
ITS sequences of Pachysandra terminalis was used as the reference sequences, we conducted the alignment comparisons to account the numbers of nucleotide transition and transversion (Table 1). The results showed that in the region of ITS-1, the numbers of bases transiting are almost similar as that of bases transve- rsing, while in the region of ITS-2 the numbers of base transiting are more than that of base transversing, this was consistent with the results of Popls plants (Shi et al., 2006). The same situation took place in the region of 5.8 S but the numbers of five or six nucleotide substitution appeared only. Little species had obvious differences reaching ten of nucleotide substitutions or more. In general, Buxus plants had much more numbers of conversions than that of transversions, and total numbers of nucleotide substitutions of Buxus plants were far greater than that of Populus plants, which indicated that nucleotide substitutions in the ITS sequences of the genus Buxus should have been significantly elevated and that nucleotide substitutions should concentrate in the ITS-1 region, accounting for 60% of the total numbers of nucleotide substitution.
 Table 1 The numbers of nucleotide substitution in ITS sequences in genus Buxus |
1.2 Nucleotide substitution rate of species in Buxus
Sequence alignment found that informative sites in ITS of Buxus genus was the uneven distribution of different intervals. Thus, we adopted the Jukes-Cantor one-parameter model and Kimura's two-parameter model to analyze the sequences in order to better reflect the real situation with ITS sequences of Pachysandra terminalis as the reference (Table 2).
 Table 2 The ratio of nucleotide substitution of ITS sequences in Buxus |
By comparison of nucleotide substitution rate, there was little differences in region of ITS-1 between one-parameter model and two-parameter model, the average difference between the two sets was 0.0107, P value of t test was 0.637 (greater than 0.05), which was not significant. Likewise, the mean difference between the two sets was 0.0063 in the region of ITS-2, P value of t test was 0.707 (greater than 0.05), indicating that the results of the two models calculated the difference should not be significant. Whereas the average difference between the two sets was 0.0004in the interval of 5.8S, P value of t test was 0.970 (greater than 0.05), which was not significant. With of whole ITS region, the average difference of the results between the one-parameter model and two-parametermodel was 0.0038, being close to the result in the region of ITS-2 due to the significant differences of conversion and transversion of nucleotide in the region of ITS-2 leading to the results from one-parameter model being less than that of the two-parameter model. So, using the two-parameter model to analyze the molecular evolution of the genus Buxus would be better corresponding to the true situation. Thus, two-parameter model was used to analyze the following results in this research. As analysis of the coefficient of variation, coefficient of variation of ITS-1 in Boxus was 12.4%, of ITS-2 was 19.3%, which the later was significantly larger than the former, indicating that variation of ITS-1 sequence was less than that of ITS-2. Whereas, coefficient of variation in 5.8 S sequence was 63.8%, which was significantly higher than that of the former two sequences. The reason might be significant changes occurring in the 5.8S sequences of three species of B. glomerate, B. gonoclada, and B. citrifolia, whose numbers of conversions and transversions were increased signifycantly, leading to variation coefficient of 5.8 S sequence increasing.
1.3 Relative rate of genus Buxus detected
First of all, using Phylip software to build phylogenetic tree of the genus Buxus, and taking Pachysandra terminalis as reference of the outgroup, the maximum parsimony tree was constructed based on the Kimura two-parameter model (Figure 1). In this study, the 18 tested plants of the genus Buxus can be divided into three major branches, B. macowanii, B. glomerata, B. gonoclada and B. citrifolia assigned to a branch. B. sempervirens, B. bodinieri and B. balearica assigned to a group; the rest assigned to a branch. These three groups were further divided into seven clustering groups, serially coded as â… , â…¡, â…¢, â…£, â…¤, â…¥ and â…¦, respectively. The relative rate was detected by using the approach proposed by Sarich and Wilson (1973), the results indicated in Table 3. The results showed that there were differences in the rate of molecular evolution of ITS sequences among the different clustering groups in Buxus. Comparative analysis of ITS-1 sequences showed that the evolution rate of groupâ… was the fastest one, with the signi- ficant differences (P <0.01) from the group â…¢ and â…£, followed by â…¢ groups and the slowest evolution rate was group â…£, with no significant differences (P <0.01) from the group â…¡, â…¢ (P> 0.05). Likewise, analysis of ITS-2 sequence showed that the fastest evolution rate was group â…°, with significant differences group â…², followed by group â…³, the slowest evolution rate was group â…², with no significant difference. Comprehensive analysis of whole ITS sequence pres- ented that the fastest evolutionary rate was groups â…°, followed by group â…², the slowest evolution rate was group â…³, all of them have reached a very significant difference.
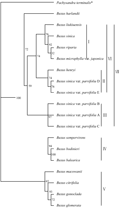 Figure 1 Phylogenetic tree of genus based on the ITS sequences |
Recently, there is yet considerable debates on the relative relationship of between pearl boxwood (Buxus sinica var. parvifolia) and Buxus sinica as well as the taxonomic status of pearl boxwood (Buxus sinica var. parvifolia). Zheng (1980) thought that the both belonged to the same plant species and further considered that pearl boxwood (Buxus sinica var. parvifolia) was a kind of the Buxus sinica’s variant. In this study we found that pearl boxwood (Buxus sinica var. parvifolia) was not a variant of Buxus sinica, but could be a kind of sister species, the rate of molecular evolution detected was different rates of evolution, the evolution rate of Buxus sinica was significantly higher than that of the pearl boxwood (Buxus sinica var. parvifolia). The karyotype analysis in our laboratory also confirmed that two species was not the same plant species, the former (Buxus sinica var. parvifolia). Would be more advanced evolutionary than the latter. In contrast, B. henryi would be much closer in the relationship than the pearl boxwood, which also had some supporting results in karyotype analysis. Ther- efore, the authors thought that pearl boxwood (Buxus sinica var. parvifolia) was not a variant of Buxus sinica, but whether pearl boxwood could be considered as being independent species, which it remains to be validated by the barcoding genes such as matK and rbcL or other genes.
There are many plant species in Buxus genus, 17 species existing in China only, and mainly distributed in the southwest of China. Therefore, Acquiring plant materials would be the difficulty to build the completely phylogenetic relationships of Buxus genus, which we will step up the work in our lab. Authors believed that, the difficulty in genus Buxus for classifying different groups might be due to the inter-species hybridization and continuity of the morphological variation. If we directly study the phylogenetic relationships of the genus at the DNA levels, it would play a great action to overcome the synonyms or homonym in genus as well as to promote the studies for domestication and breeding of Buxus plant, biodiversity research and applications in garde- ning and landscape.
Molecular systematics is to study the history of biolo- gical evolution and reconstruction of phylogenetic relationships based on the information of biological macromolecules, of which the theoretical basis, known as the “molecular clock hypothesis”, is the evolutionary rate is relatively constant with biological macromolecules in the evolutionary events (Tang et al., 2002). However, since the hypothesis has been proposed, there were more contentious in the studies of phylogenetics in higher organisms (Muse, 2000), especially, whether the rate of molecular evolution in different cluster groups of the same organisms is relatively consistent. Many scholars have long suspe- cted that this hypothesis be reasonable, and some experimental evidenceshas been come out negative, for examples, Hartl and Clark (1997) and Vawter and Brown (1986) independently proposed that different gene should have different evolutionary rate. Zhang and Ryder (1993) found the same gene should be significantly differences in evolutionary rate in different biological groups. Therefore, the same gene with allometric phenomena of molecular evolution would be one of the major challenges for molecular clock hypothesis (Luo and Zhang, 2000). In this study, The results showed that there was a greater difference in the rate of nucleotide substitution of ITS gene in the genus Buxus as well as a significant difference in the rate of evolution among cluster groups, so this would provide new evidence not only for the molecular allometric phenomenon in the evolutionary process, but also for in-depth understanding phylogenetic relationships the genus Buxus.
3 Materials and Methods
3.1 Experimental materials
In this study, five species of Buxus genus widely distributed in China, including five populations of pearl boxwood (four wild species, one cultivated species) were used for molecular evolution studies. In addition, nine ITS sequences of Buxus plants were downloaded from GenBank, and reference sequence was Pachysandra terminalis that has close genetic relationship with the Buxus plants (Table 4).
3.2 DNA extraction
Using a modified CTAB method we extracted genomic DNA of Buxus species following with the reference of Lv et al (2007).
3.3 ITS sequences amplified by PCR
Using 50 μL PCR reaction system including the double-tranded DNA template 50~100 ng, MgCl2 (25 mmol/L), 4 μL, dNTP (2.5 mmol/L), 6 μL, Taq DNA polymerase (TaKaRa) 1.75 U, forward primer 0.75 μL (5'-AGAAGTCGTAACAAGGTTTCCGTAG G-3', reverse primer 20 μmol/L, 5'-TCCTCCGCT TATTGATATG C-3', 20 μmol/L), and adding ddH2O to 50 μL. PCR amplification procedures as 94℃ pre-denaturation 4 min, and then 30 cycles with 94℃ denaturation for 30 s, 57℃ annealing for 30 s, and 72℃ extension for 1 min, finally, after 72℃ extension 7 min, placed at 4℃ ready for use. PCR products were purified by using purification kit (AxyPrep DNA Gel Extraction Kit) to be as sequencing template for sequencing directly.
3.4 ITS sequencing
Sequencing was performed by Shanghai Yinjun Biotechnology Company. Adopting two of four classic sequencing primers in the 5.8S reported previously, P2: (5'-GCATCGATGAAGAACGCAGC-3') and P3 (5'-GCTGCGTTCTTCATCGATGC-3') (Wang et al., 1999) for the sequencing primers, synthesized by Shanghai Yinjun Biotechnology Company.
 Table 3 The molecular evolution relative rate of ITS sequences in different clusters of Buxus genus |
 Table 4 The materials used in the research and their sources |
3.5 Statistical Methods
Using ITS sequence of Pachysandra terminalis as a reference, the number of nucleotide substitutions was counted out based on the principle of using pair-wise comparison. Using one parameter model of Jukes and Cantor (1969) and two-parameter model of Kimura (1968) calculated the base substitution data.
Authors’ Contributions
XFW and NL were the persons who conducted experimental design and performances and XFW also finished the data analysis and wrote the manuscript. KSJ was principal investi- gator who conceived the project and supervised the experimental designing, data analyzing, and paper writing and revising. All authors have read and agreed the final context of manuscript.
Acknowledgement
This study was funded by the “Eleventh Five-Year” National Science and Technology Support Project (NO.2006BAD01A1403). The authors thanked Zhong- shan Botanical Garden of Chinese Academy of Sciences for providing plant materials.
References
Hartl D.L., and Clark A.G., 1997, Principles of population genetics (third edition), Sinauer Associates, Inc. Publishers, Sunderland, Mass- achusetts, USA, pp.1-542
Huang Y., Ji K.S., Jiang Z.H., and Tang G.G., 2008, Genetic structure of Buxus sinica var. parvifolia, a rare and endangered plant, Scientia Horticul- turae, 116(3): 324-329
http://dx.doi.org/10.1016/j.scienta.2008.01.001
Jiang Z.H., Ji K.S., and Huang Y., 2008, Analysis of genetic diversity of two populations of Buxus sinica var. parvifolia by RAPD, Nanjing Linye Daxue Xuebao (Journal of Nanjing Forestry University), 32(1): 11-14
Jukes T.H., and Cantor C.R., 1969, Evolution of protein molecules, In: Munro H.N. (ed.), Mam- malian Protein Metabolism, Academic Press, New York, pp.21-132
Kimura M., 1968, Evolutionary rate at the molecular lever, Nature, 217: 624-626
http://dx.doi.org/10.1038/217624a0 PMid:5637732
Lin Q., 2004, A new synonym of Buxus rugulosa Hatusima, Zhiwu Yanjiu (Bulletin of Botanical Research), 24 (4): 402-403
Luo J., and Zhang Y.P., 2000, Molecular clock and its question, Renlei Xue Xuebao (Acta Anthro- pologica sinica), 19(2): 151-159
Lv L.Y., and Ji K.S., 2009, Identification on clones of Buxus sinica var. parvifolia by ISSR, Linye Keji Kaifa (China Forestry Science and Technology), 23(5): 87-89
Lv L.Y., and Ji K.S., 2009, Peroxidase isozyme study of Buxus sinica var. parvifolia clones, Anhui Nongye Kexue (Journal of Anhui Agri. Sci.), 37(30): 14600-14601
Lv L.Y., Ji K.S., and Huang Y., 2007, Comparison of genomic DNA extraction methods and optimi- zation of ISSR-PCR reaction systems for Buxus sinica var. parvifolia, Fenzi Zhiwu Yuzhong (Molecular Plant Breeding), 5(1): 145-150
Muse S.V., 2000, Examining rates and patterns of nucleotide substitution in plants, plant molecular biology, 42(1): 25-43
Sarich V.M., and Wilson A.C., 1973, Generation time and genomic evolution in primates, Sci., 179(78): 1144-1147
http://dx.doi.org/10.1126/science.179.4078.1144 PMid:4120260
Shi Q.L., Zhu G.Q., Huang M.R., and Wang M.X., 2006, The Characteristic of Molecular Evolution in Poplus based on ITS Sequence Analysis, Fenzi Zhiwu Yuzhong (Molecular Plant Breeding), 4(2): 255-261
Tang X.H., Lai X.L., Zhong Y., Li T., and Yang S.J., 2002, Hypothesis of molecular clock and fossil record, Dixue Qianyuan (Earth Science Frontiers), 9(2): 465-474
Vawter L., and Brown W.M., 1986, Nuclear and mitochondrial DNA comparisons reveal extreme rate variation in the molecular clock, Science, 234(4473): 194-196
http://dx.doi.org/10.1126/science.3018931 PMid:3018931
Wang J.B., Zhang W.J., and Chen J.K., 1999, Application of ITS sequences of nuclear rDNA in phylogenetic and evolutionary studies of angiosperms, Zhiwu Fenlei Xuebao (Acta Phytotaxonomica Sinica), 37(4): 407-416
Wang L.L., Xu N., Li J.Y., and Guan Y., 2008, A study on the esterase and peroxidase isozymes of several species in Buxus, Harbin Shifan Daxue Ziran Kexue Xuebao (Natural Sciences Journal of Harbin Normal University), 24(6): 74-76
Yan S.X., Zhao Y., and Zhao T.B., 2002, A quantitative classification stady on the plants of Buxus L. from China, Shengwu Shuxue Xuebao (Journal of Biomathematics), 17(3): 380-383
Zhang Y.P., and Ryder A.O., 1993, Mitochondrial DNA sequence evolution in the arctoidea, Proc. Natl. Acard. Sci. USA, 90(20): 9557-9561
http://dx.doi.org/10.1073/pnas.90.20.9557
Zheng M., ed., 1980, Buxaceae, flora reipublicae popularis sinicae, Science Press, Beijing, China, pp.16-60
. PDF(203KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Xiaofeng Wang
. Nana Liu
. Kongshu Ji
Related articles
. Buxus spp
. Chinese pearl boxwood
. B. sinica var. parvifolia
. ITS sequence
. Molecular evolution
. Nucleotide substitution
Tools
. Email to a friend
. Post a comment


