2. Rice Research Institute, Jiangxi Academy of Agricultural Sciences, Nanchang, 330200, P.R. China
 Author
Author  Correspondence author
Correspondence author
Plant Gene and Trait, 2011, Vol. 2, No. 4 doi: 10.5376/pgt.2011.02.0004
Received: 10 Oct., 2011 Accepted: 11 Dec., 2011 Published: 22 Jan., 2012
He et al., 2011, Genetic Analysis and Gene Mapping of a Rice White Stripe Leaf Mutant (st10), Plant Gene and Trait, Vol.2, No.4 23-29 (doi: 10.5376/pgt.2011.02.0004)
A temperature-sensitive white-stripe leaf mutant st10 was identified from rice mutant library generated by EMS mutagenesis in our previous work, which has a genomic background of elite japonica variety Nipponbare. The leaves of mutant st10 appears white-stripe fore and aft in two or three leaf stages, then the white color gradually weakens as the plant grows, leaf color returns to normal during heading stage, besides the leaf vein continuing to show white color. The white strip trait is obviously affected by temperature change. All of mutants would well exhibit their mutant traits growing under the high temperature condition, which appear white strip leaves and even full white panicles, whereas the white-stripe of leave becomes narrow and the color of panicles appears normal growing the low temperature condition. The results of genetic analysis revealed that white-stripe phenotype was controlled by a single recessive nucleic gene. The F2 population was developed from a across between peiai64s (a male sterility line) and st10. st10 gene was mapped in 150 kb region between marker STR19 and marker STR24 on chromosome 3. In this research we assayed the chlorophyll contents of mutant leaves growing under the temperatures at 24℃, 28℃ and 32℃, the result showed that the content of chlorophyll b and chlorophyll a were significantly reduced at the temperature of 32℃, while there is no obviously change at 24℃or at 28℃. Chlorophyll Fluorescence data revealed that maximum photosynthetic quantum capacity of the mutant was less than that of the wild-type, which would contribute the decrease of chlorophyll contents of the mutant. In this study, we constructed the genetic separate population by using Pei-Ai 64, an elite sterile line, as maternal parent and st10 mutant as paternal parent to further analyze the genetic basis, the result demonstrated that the phenotype of the mutant trait was controlled with a single recessive locus that was mapped on chromosome 3 within the 150 kb region framed between STR19 and STR 24.
Chlorophyll affects crop photosynthetic efficiency through involving in capturing the light energy, charge separation and electron transfer in antenna complexes (Tanaka and Tanaka, 2006). The current study has shown that leaf color mutant gene could be directly or indirectly affect the synthesis and degradation of chlorophyll to alter chlorophyll content of mutant, consequently leading to decrease of photosynthetic efficiency to reduce crop yield and even to result in plant death (He et al., 2006). Leaf color mutations can be divided into many different types according to the colors of seedling leaves, such as yellow, albino, light green, stripes, zebra, etc. (Wu, 1995). The studies of plant leaf color mutations have great significances to in-depth understand the relationships of chlorophyll synthesis and degradation pathways as well as to understand plant photosynthesis to further explore the mechanism of plant photosynthesis.
In recent years, it has been reported on mutations of the uneven leaf color in rice, such as the white stripe leaf, zebra leaves and so on, and most of the reported mutants were associated with temperature, the mutant phenotypes were different under the different temperature conditions. Therefore, the studies of temperature-sensitive leaf color mutation will help to further clarify the mechanism of leaf color change, which might be the basis for in-depth exploring the relationship between photosynthesis and temperature response. Xie et al found leaf color mutant 11035 from the rice field population of TGMS 89025, a Photo-thermoperiod Sensitive Genic Male Sterile Lines, in which is characterized by the changes of intermittent loss of green happening in leaves and leaf sheaths in the low temperature whether in the seedling stage, the "zebra-type stripes" with permutation of yellow and white in whole plant occurred prior to recovering normal green as temperature had gone up again in a few days later (Xie et al., 1995). Genetic analysis of the zebra leaf B411 mutant showed the mutant trait was controlled by one pair of recessive gene. Chloroplast ultrastructure observations of yellow zone and green zone of zebra leaf revealed that development of chloroplast in green zone of zebra leaf was normal, while the shape of the chloroplast in the yellow zone was abnormal, and inner-membrane systems of the chloroplast was also incomplete, the chloroplast contained osmiophilic bodies, with the yellow zone gradually re-greening, normal chloroplast structure will being recovered (Li et al., 2008). Rice zebra leaf gene, ZEBRA2, encodes carotenoid isomerase, of which the key is the enzyme to convert cis-lycopene into trans-lycopene, mainly expressed in mesophyll cells of mature leaves, involving in the process of photo-protection. Phenotype of zebra leaf might be related to light oxidative damage in strong light conditions (Chai et al., 2010). SooCheul Yoo et al (2009) reported a lethal mutant of tobacco leaf zebra zn (zebra-necrosis), which the phenotype is yellow and green zebra stripes occuring in the leaf sheaths cultured in high and low alternating temperatures before the leaves grown up. The gene encodes an unknown functional thylakoid-binding protein, which might be paly important roles in the accumulation process of the thylakoid protein complexes from dark to light conditions (Li et al., 2010). And Soo-Cheul Yoo et al also found that white leaf stripe mutant genes st1 (Strip1) encodes the small subunit of ribonucleotide reductase, which involves chloroplast formation during the leaf growth and development. The leaves of mutant st1 appears white stripes under the continued temperatures of 20℃ or 30℃, while cultured at alternative changes of 20℃ and 30℃ daily, the leaves will turn color normal (Soo-Cheul et al., 2009). Xu et al (2010) found that a temperature-sensitive white stripe mutant of rice was allelic to white stripes st1, but the white-striped phenotype of a temperature-sensitive white stripe mutant appears after the second leaf growing, while the white stripes of st1 appears after the 4th or 5th leaf growing.
We have identified a temperature-sensitive white stripe leaf mutant with the background of japonica Nipponbare from the EMS mutant library constructed in our lab. Genetic analysis showed that the mutant trait was controlled by a pair of recessive nuclear genes. In this study, the chlorophyll content, chlorophyll fluorescence parameters and various agronomic traits were measured and analyzed. Genetic mapping was carried out by using separating mapping population derive from the cross of PeiAi 64 s and mutant st10. Finally, the target gene was mapped on the short arm of chromosome 3 framed between the markers of STR19 and STR24 in the range 150 kb, which laid basis for gene cloning and in-depth study of the functions.
1 Results
1.1 Phenotypic and genetic analysis
White stipe leaves of the mutant appeared in the 2 or 3 leaf stage as shown in Figure 1. The whole leaf of some plants happened in whiteness or even the whole plant become whitening. As the plant grew white stripe in the leaves become narrow, some white stripes in the leaf disappeared and leaves stayed with light color and pale vein compering to the wild type. Leaf color of mutant was sensitive to the change of the temperature; white stripe will be moreobvious in high-temperature than in low-temperature. As shown in Figure 1A, the phenomenon of white panicle occurred in the high temperature while the color of panicle was normal in low temperature. During the September 2010, Hangzhou, consecutive hot weather happed, panicle of mutant whitened, the contents of chlorophyll a and chlorophyll b reduced, in which content of chlorophyll a was 88% content of Nipponbare while the content of chlorophyll b was 92% of the content of Nipponbare. The numbers of tillers little change between the wild-type and the mutant, whereas plant height of the mutant was significantly shorter, stems became thinner, Kilo-seed weight decreased by 10%, spike length and tiller number did not significantly change any more (Table 1). We employed 9311 as paternal parent and mutant st10 as maternal parent as well as Peiai 64s as maternal parent and mutant st10 paternal parent, respectively, to make crosses, the F1 plants exhibited normal phenotype, the phenotypic separation of F2 individuals in the population occurred, of which the normal phenotype of plants to mutant phenotype was close to 3 to 1, the Chi test results (χ2=0.139 <χ20.05=3.84; χ2=0.769<χ20.05=3.84) (Table 2) indicated that the mutant trait would be controlled by a pair of single recessive nuclear gene.
 Figure 1 Phenotypes of wild-type Nipponbare, and the st10 mutant at different growing stages |
Table 1 Phenotypic characteristic of the mutant st10 and the wild-type Nipponbare |
Table 2 Genetic analysis of mutant st10 |
1.2 erature gradient experiments
In order to clarify the critical temperature of mutant in the temperature-sensitive change, the experiment of mutant st10 and wild-type Nipponbare were treated to be growing with the same growth conditions except the temperatures at 24℃, 28℃ and 32℃, respectively. The phenotype of the mutant was more obvious at 32℃ than at 24℃ or 28℃. The chlorophyll of mutant and wild-type was not significantly differences in temperature treatments at 24℃ or 28℃. The leaf color of mutant was turning clear whiteness at 32℃ and chlorophyll content was also decreasing of which content of chlorophyll a was decreasing by 17%, as well as decrease of chlorophyll b by 11%. The chlorophyll content under the three temperature conditions at three-leaf stage were shown in Figure 2.
Figure 2 Pigment content of st10 at the treatments of different temperatures |
1.3 nalysis of green fluorescence
To further analyze mutant photosynthetic potential, we also measured the chlorophyll fluorescence paraters of Nipponbare and the mutant. The photochemical efficiency of PS â…¡ refers to the number of the transmitting electron or the time of electronic charge separation occurs in reaction center while PSâ…¡ absorbs a photon, whi`h usually presented with chlorophyll fluorescence parameters Fo', Fm, FV/Fm (Shen et al., 2009). Fo', Fv/Fm', Fm' are the three parameters under the conditions of light reaction, presenting the status of light reaction of sample.
As indicated in Figure 3, under the light reaction conditions, three parameters of mutant were smaller than that of wild-type, the FV/Fm' of mutant st10 declined 13% off compared to Nipponbare, indicating yield of mutant st10 was slightly lower than that of Nipponbare leading to reduce photosynthetic efficiency of PS â…¡ of the mutant. Under the dark reaction conditions, the F0 of mutant increased by 7% than that of wild-type, whereas the Fv/Fm of mutant decreased by 17% off than that of wild-type (Figure 4). Therefore, photosynthetic efficiency of the mutant decreased more obvious under the dark reaction conditions than under the light reaction conditions. The dark-adapted conditions, the amount of maximum fluorescence and minimum fluorescence of the mutant was significantly less than that of the wild type. As the mutant captured less the amount of light energy in the carbon assimilation process of photosynthesis, the amount of CO2 fixed in primary reaction also decreased, thereby the plant photosynthetic efficiency reduced. The chlorophyll contents of mutant and Nipponbare were quite similar in tillering stage, so the photosynthetic electron transferring process can still be normal maintained.
.png) Figure 3 Pigment content of st10 at the treatments of different temperatures |
.png) Figure 4 Pigment content of st10 at the treatments of different temperatures |
1.4 rimary mapping and fine mapping of st10 gene
We used published SSR markers evenly distributed in rice genome to scan the polymorphic SSR markers between the genomes of Nipponbare and PeiAi 64s. The 64 pairs of SSRs with polymorphisms were obtained for primary mapping in this research. Thirty of randomly selected seedlings with mutant phenotype in the F2 population were mixed to extract DNA as a mixed DNA pool. The mutant gene was primary mapped on chromosome 3 framed between CHL-8 and RM16 with the genetic distances of 86.0 cM and 91.1 cM, respectively. In order to further fine mapping the target gene, we searched the markers in this region and designed 19 pairs of STS (Sequence Tagged Site) markers, polymorphic analysis showed that 10 of the STS markers between the two parents exhibited significantly polymorphisms (the marker sequence shown in Table 3). By using 2885 individuals of F2 mapping populations, the mutant gene was finally mapped on chromosome 3 framed between the STR19 and STR24 in the range of physical distance about 150 Kb, 19 exchanging plants in the left flanking occurring, while 3 exchanging plants in the right flanking (Figure 5, Figure 6).
Table 3 Markers used for fine mapping of st10 gene |
Figure 5 Electrophoretic banding pattern generated by STR6 marker for fine mapping among parts of plants with st10 phenotype in F2 population |
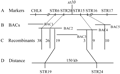 Figure 6 Fine mapping of st10 on rice chromosome 3 |
2 Discussions
Temperature-sensitive leaf color mutant is the ideal material to study the biochemical processes and biological origin of chloroplasts in higher plants as well as to clarity the mechanism of leaf color changes, also to in-depth explore the relationship between photosynthesis and temperature response. Liu et al. found a yellow-green leaf mutant of rice, which the leaf color was normal when the temperature was below 23℃, while yellow-green phenotype of leaves occurred once the temperature was higher than 26℃. With the rise of the temperature chlorophyll content of mutant leaves decreased in trends. At the conditions of low temperatures chlorophyll content of the mutant and wild-type were quite similar, whereas at the conditions of high temperature, the contents of chlorophyll a and chlorophyll b of the mutant was significantly lower than that of the control. Under field conditions, the chlorophyll a content of the mutant in seedling stage decreased by 30% compared with wild-type and the content of chlorophyll b decreased by 50%. The gene was identified as Cde1(t), encoding a glutamyl-tRNA synthetase, that was the key enzyme to catalytic bind glutamic acid and t-RNA, playing an important role in the ALA (5-aminolevulinic acid) synthesis (Liu et al., 2007). Dong et al studies on rice heat-sensitive leaf mutant (tsc1) 7436S showed that mutant phenotypes presented three colors of white, light green and green at the temperatures of 23.1℃, 26.1℃ and 30.1℃ , respectively. However, the seedlings treated at temperature 23.1℃ were transferred to the normal temperature of 30.1℃, the mutant phenotype recovered to normal green color. Transmitting electronic microscopic (TEM) analysis showed that thylakoid structure of mutant was abnormal bubbly-like and grana lamellae structure disappeared compared with the control (Dong et al., 2001). Wu et al found a green turning type albino mutant W25 that the leaf color was completely white-ning in temperature conditions of 15℃, 20℃ and 25℃, while at 30℃ and 35℃, the first or second leaf exhibited light green or normal green (Wu et al., 1996). Meanwhile, temperature-sensitive mutants had been reported in other species, such as in Arabidopsis (Markwell et al., 1992), cotton (Alberte et al., 1974), barley (Gálová et al., 2000), soybean (Stockinger et al., 1994), corn (Pasini et al., 2005).
In recent years, it has been many reports on the studies of mutations of uneven color leaves in rice (such as white stripe leaves, zebra leaves, etc.) , however, these reports were mostly concentrated on the studies of in the aspects of morphology and physiology of the mutants, it remains to be clarified the formation mechanism of mutant and regulating gene, mechanism of occurrence in the similarities and differences of leaf color uneven or uniform mutation, and influences of the temperature on the mutant phenotype. In this study, the characteristics of temperature-sensitive white stripe mutant st10 were determined including chlorophyll fluorescence parameters, agronomic traits and chlorophyll, while the mutant gene was fine mapped on chromosome 3 within the range of 150 Kb, which might lay fundaments for exploring the molecular mechanism of leaf color mutation and regulation of temperature on the gene expression related to leaf color mutation in plant.
3 Materials and Methods
3.1 Materials used in this study
Mutants st10 used in our laboratory was obtained from the mutagenesis of japonica variety Nipponbare induced with ethyl methane sulphonate (EMS). F2 segregating population as mapping population was derived from the cross between maternal parent Peiai 64s and paternal parent st10 mutant constructed.
3.2 Determination of chlorophyll content
Taking flag leaves from the mutant and the control Nipponbare in tillering stage and removing the main vein, cutting small pieces in side length of 2~3 mm. Taking leaf pieces of 0.15~0.2 g soaked in 25 mL in a mixture of acetone and ethanol (95% acetone: ethanol=2:1) with 4 replicates dark cultured at 26℃ for 24 hours. Taking the solution to measure the optical density values of chlorophyll a, chlorophyll b and carotenoids in the UV spectrophotometer (DU640) with three wavelengths of 470 nm, 645 nm and 663 nm. The contents of chlorophyll a, chlorophyll b and carotenoid were calculated following with Arnon's method (Arnon, 1949).
3.3 Temperature gradient experiment
Germinated seeds of the mutant and Nipponbare were placed on media with 0.8% agar and cultured with lighting for 24 hours at three temperatures of 24℃, 28℃ and 32℃, respectively. Observing and recording the mutant’s phenotype, taking leaves to determine the content of chlorophyll until the mutant phenotype is obviously exhibiting in three leaf stage.
3.4 Determination of chlorophyll fluorescence parameters
Using a portable chlorophyll fluorometer (PAM-2500, Germany) measured values of rice flag leaf under the light reaction conditions as well as the values of Fo, Fm and Fv/Fm under the dark reaction conditions. The experiment was performed at 9:00-10:00 am on July 23, 2010 (in tillering stage) with three replicates, the averaged measure values were used for calculation following with the formula as below:
Fv/Fm=(Fm-Fo)/Fm
[Fv'/Fm'=(Fm'-Fo')/Fm'
Determination in light reaction: when the measured leaves being adapted to light, turning on the measuring light to determine steady-state fluorescence of the leaves, and then turning on the saturation pulse light to measure the maximum fluorescence Fm', turning off the actinic light and turning on far-red light to excite PS â… making the PS â…¡ electron transporters in the status of oxidation prior to measuring the minimum fluorescence Fo' of light-adapted leaves.
Determination in dark reaction: Using the dark-adapted leaf folder to hold rice flag leaf on the position from one third base, closing the folder and staying 20 min in dark environment. Putting the light-emitting diodes into the hole of the leaf folder, turning on the measuring light to obtain Fo value, then turning the saturation pulse to obtain Fm values, subsequently turning the action light,1 min later turning the satu-ration pulse to measure Fv/Fm (Bo Shen et al., 2009).
3.5 Molecular tagging of mutant st10
Map-based cloning approach was employed in this study. We used published SSR markers to scan the genome-wide polymorphic SSR markers between the genomes of Nipponbare and PeiAi 64s. The scanned 64 pair SSRs with polymorphisms was used for primary mapping. Ten STS markers with obvious polymorphisms were developed by using the softwares of Premier 5.0 and DNAMAN of which primers were synthesized by Shanghai Yingwei Jieji Company. Physical locations on chromosome were following the NCBI website of http://www.ncbi.nlm.him.nih.gov. PCR reaction performed as follows: in total of 20 μL reaction volumes including: DNA template 2 μL; 10 × PCR buffer 2 μL; dNTP (2.5 mmol/L) 2 μL; Primer F (2.5 μmol/L) 2 μL; Primer R (2.5 μmol/L) 2 μL; Taq polymerase 1 μL; adding ddH2O 9 μL up to 20 μL. Reaction procedures as follows: 94℃ pre-denaturing 4 min, then 40 cycles of 94℃ denaturing 40 s, annealing 55℃ 40 s, 72℃ extending 40 s, finally 72℃ extending 10 min, keeping at 15℃ ready for use. Amplified products were separated with 4% or 5% agarose gel electrophoresis, stained by ethidium bromide and observed under UV lamp.
Author’s contributions
HYH GXZ and YCR are the persons who carried out this experiment; JH, JL, ZYG and LBG participated in some in lab work; and the data analysis and in field work; Ljand OQ conceived the project and designed the experiments as well as wrote and revised manuscript. All authors had read and agreed the final text.
Acknowledgements
This research is jointly sponsored by the project of Zhejiang Foundation of Natural Sciences (Y3100357), National Transgenic Variety Breeding Project (2008ZX08001-002), National 973 Project (No. 2009CB118500) and project of National foundation of Natural Sciences (No. 30871513). Authors thank for two anonymous reviewers with their critical comments. In this paper we mentioned some chemical and reagent suppliers and sequencing service providers, that doesn't mean we would like to recommend or endorse the production of theirs.
References
Alberte R.S., Hesketh J.D., Hofstra G., Thornber J.P., Naylor A.W., Bernard R.L., Brim C., Endrizzi J., and Kohel R.J., 1974, Composition and activity of the photosynthetic apparatus in temperature-sensitive mutants of higher plants, Proc. Natl. Acad. Sci., USA, 71: 2414-2418
http://dx.doi.org/10.1073/pnas.71.6.2414
Arnon D.I., 1949, Copper enzymes in isolated chloroplasts: Polyphe-noloxidase in Beta vulgaris, Plant Physiol., 24: 1-15
http://dx.doi.org/10.1104/pp.24.1.1 PMid:16654194 PMCid:437905
Chai C.L., Fang J., Liu Y., Tong H.N., Gong Y.Q., Wang Y.Q., Liu M., Wang Y.P., Qian Q., Cheng Z.K., and Chu C.C., 2010, ZEBRA2, encoding a carotenoid isomerase, is involved in photo protection in rice, Plant Mol. Biol., 1573-5028
Dong Y.J., Dong W.Q., and Shi S.Y., 2001, Identification and genetic analysis of a thermo sensitive seeding colour mutant in rice, Breeding Science, 51: 1-4
http://dx.doi.org/10.1270/jsbbs.51.1
Gálová E., Bhmová B., and Svoviová A., 2000, Analysis of some barley chlorophyll mutants and their response to temperature stress, Photosynthetica, 38: 29-35
http://dx.doi.org/10.1023/A:1026735605804
He B., Liu L.L., Zhang W.W., and Wan J.M., 2006, Plant leaf color mutants, Zhiwu Shenglixue Tongxun (Plant Physiology Communications), 42(1): 1-9
Li H., 2008, The Expression conditions and inheritance of zebra-leaf trait in rice, Thesis for M.S., Hunan Normal University, Supervisors: Chen L.B., and Qiu Y.L., pp.1-48
Li J., Pandeya D., Nath K., Zulfugarov I.S., Yoo S.C., Zhang H., Yoo J.H., Cho S.H., Koh H.J., Kim D.S., Seo H.S., Kang B.C., Lee C.H., and Paek N.C., 2010, ZEBRA-NECROSIS, a thylakoid-bound protein, is critical for the photoprotection of developing chloroplasts during early leaf development, Plant J., 62: 713-725
http://dx.doi.org/10.1111/j.1365-313X.2010.04183.x PMid:20202171
Liu W., Fu Y.P., Hu G.C., Si H.M., Zhu L., Wu C., and Sun Z.X., 2007, Identification and fine mapping of a thermosensitive chlorophyll deficient mutant in rice (Oryza sativa L.), Planta, 226: 785-795
http://dx.doi.org/10.1007/s00425-007-0525-z PMid:17541632
Markwell J., and Osterman J.C., 1992, Occurrence of temperature-sensitive phenotypic plasticity in chlorophyll deficient mutants of Arabidopsis thaliana, Plant Physiol., 98: 392-394
http://dx.doi.org/10.1104/pp.98.1.392 PMid: 16668642 PMCid:1080195
Pasini L., Bruschini S., Bertoli A., Mazza R., Fracheboud Y., and Marocco A., 2005, Photosynthetic performance of cold-sensitive mutants of maize at low temperature, Physiol. Plant, 124: 362-370
http://dx.doi.org/10.1111/j.1399-3054.2005.00522.x
Shen B., Jiang L., Yu W.D., Fan Y.Y., and Zhuang J.Y., 2009, QTL Analysis of chlorophyll fluorescence parameters in rice seedlings under salt stress, Zhongguo Shuidao Kexue (China J. Rice Sci.), 23(3): 319-322
Soo-Cheul Y., Sung-Hwan C., Hiroki S., Jinjie L., Kensuke K., Hee-Jong K., Koh I., and Nam-Chon P., 2009, Rice virescent3 and stripe1 encoding the large and small subunits of ribonucleotide reductase are required for chloroplast biogenesis during early leaf development, Plant Physiology, 150: 388-401
http://dx.doi.org/10.1104/pp.109.136648 PMid:19297585 PMCid:2675711
Stockinger E.J., and Walling L.L., 1994, A chlorophyll a/b-binding protein gene from soybean, Plant Physiol., 104: 1475-1476
http://dx.doi.org/10.1104/pp.104.4.1475 PMid:8016279 PMCid:159320
Tanaka A., and Tanaka R., 2006, Chlorophyll metabolism, Curr. Opin. Plant Biol., 9: 248-255
http://dx.doi.org/10.1016/j.pbi.2006.03.011 PMid:16603411
Wu D.X., Shu Q.Y., Xia Y.W., Zheng T., and Liu G.F., 1996, Leaf color character and genetics of a new greenable albino mutation line W25 of rice, 1996, Zhejiang Nongye Xuebao (Acta Agriculturae Zhejiangensis), 8: 372-374
Wu D.X., Xia Y.W., and Shu Q.R., 1995, The research and application of Chlorophyll mutant plants, Zhongguo Nongxue Tongbao (Chinese Agricultural Science Bulletin), 04: 36-39
Xie R., Zhu F.Y., He G.H., Deng X.H., Zuo Y.S., Yang Z.L., and Wu L.J., 1995, Preliminary research in natural mutant of thermo-sensitive chlorophyll in two-line rice, Xinan Nongye Xuebao (Southwest China Journal of Agricultural Sciences), 8(A01): 124-128
Xu F.H., Cheng Z.J., Wang J.L., Wu Z.M., Sun W., Zhang X., Lei C.L., Wang J., Wu F.Q., Guo X.P., Liu L.L., and Wan J.M., 2010, Genetic analysis and fine-mapping of Gws gene using green-white-stripe rice mutant, Zuo wu Xue bao (Acta Agronomica Sinica), 05: 713-720
http://dx.doi.org/10.3724/SP.J.1006.2010.00713
. PDF(298KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Yinghong He
. Guoxing Zou
. Yuchun Rao
. Jiang Hu
. Jian Liu
. Zhenyu Gao
. Longbiao Guo
. Li Zhu
. Qian Qian
Related articles
. Rice mutant ( Oryza sativa L. spp japonica )
. White-stripe leaf
. Genetic analysis
. Molecular mapping
Tools
. Email to a friend
. Post a comment


